Expansion microscopy: development and neuroscience applications
- PMID: 29316506
- PMCID: PMC5984670
- DOI: 10.1016/j.conb.2017.12.012
Expansion microscopy: development and neuroscience applications
Abstract
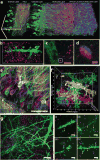
Many neuroscience questions center around understanding how the molecules and wiring in neural circuits mechanistically yield behavioral functions, or go awry in disease states. However, mapping the molecules and wiring of neurons across the large scales of neural circuits has posed a great challenge. We recently developed expansion microscopy (ExM), a process in which we physically magnify biological specimens such as brain circuits. We synthesize throughout preserved brain specimens a dense, even mesh of a swellable polymer such as sodium polyacrylate, anchoring key biomolecules such as proteins and nucleic acids to the polymer. After mechanical homogenization of the specimen-polymer composite, we add water, and the polymer swells, pulling biomolecules apart. Due to the larger separation between molecules, ordinary microscopes can then perform nanoscale resolution imaging. We here review the ExM technology as well as applications to the mapping of synapses, cells, and circuits, including deployment in species such as Drosophila, mouse, non-human primate, and human.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
-
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, Lichtman JW. Saturated Reconstruction of a Volume of Neocortex. Cell. 2015;162(3):648–661. doi: 10.1016/j.cell.2015.06.054. - DOI - PubMed
-
- Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, Schneider-Mizell CM, Saumweber T, Huser A, Eschbach C, Gerber B, Fetter RD, Truman JW, Priebe CE, Abbott LF, Thum AS, Zlatic M, Cardona A. The complete connectome of a learning and memory centre in an insect brain. Nature. 2017;548(7666):175–182. doi: 10.1038/nature23455. - DOI - PMC - PubMed
-
- He J, Zhou R, Wu Z, Carrasco MA, Kurshan PT, Farley JE, Simon DJ, Wang G, Han B, Hao J, Heller E, Freeman MR, Shen K, Maniatis T, Tessier-Lavigne M, Zhuang X. Prevalent presence of periodic actin–spectrin-based membrane skeleton in a broad range of neuronal cell types and animal species. Proc Natl Acad Sci. 2016;113(21):6029–6034. doi: 10.1073/pnas.1605707113. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

