Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism
- PMID: 29317493
- PMCID: PMC5846160
- DOI: 10.1074/jbc.M117.810101
Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism
Abstract
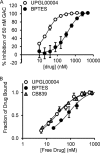
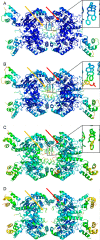
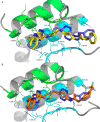
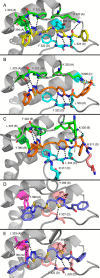
Altered glycolytic flux in cancer cells (the "Warburg effect") causes their proliferation to rely upon elevated glutamine metabolism ("glutamine addiction"). This requirement is met by the overexpression of glutaminase C (GAC), which catalyzes the first step in glutamine metabolism and therefore represents a potential therapeutic target. The small molecule CB-839 was reported to be more potent than other allosteric GAC inhibitors, including the parent compound bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl (BPTES), and is in clinical trials. Recently, we described the synthesis of BPTES analogs having distinct saturated heterocyclic cores as a replacement for the flexible chain moiety, with improved microsomal stability relative to CB-839 and BPTES. Here, we show that one of these new compounds, UPGL00004, like CB-839, more potently inhibits the enzymatic activity of GAC, compared with BPTES. We also compare the abilities of UPGL00004, CB-839, and BPTES to directly bind to recombinant GAC and demonstrate that UPGL00004 has a similar binding affinity as CB-839 for GAC. We also show that UPGL00004 potently inhibits the growth of triple-negative breast cancer cells, as well as tumor growth when combined with the anti-vascular endothelial growth factor antibody bevacizumab. Finally, we compare the X-ray crystal structures for UPGL00004 and CB-839 bound to GAC, verifying that UPGL00004 occupies the same binding site as CB-839 or BPTES and that all three inhibitors regulate the enzymatic activity of GAC via a similar allosteric mechanism. These results provide insights regarding the potency of these inhibitors that will be useful in designing novel small-molecules that target a key enzyme in cancer cell metabolism.
Keywords: BPTES; CB-839; anticancer drug; cancer; crystallography; glutaminase; metabolism.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

