Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP
- PMID: 29317501
- PMCID: PMC5827432
- DOI: 10.1074/jbc.RA117.000477
Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP
Abstract
The accumulation of misfolded proteins promotes protein aggregation and neuronal death in many neurodegenerative diseases. To counteract misfolded protein accumulation, neurons have pathways that recognize and refold or degrade aggregation-prone proteins. One U-box-containing E3 ligase, C terminus of Hsc70-interacting protein (CHIP), plays a key role in this process, targeting misfolded proteins for proteasomal degradation. CHIP plays a protective role in mouse models of neurodegenerative disease, and in humans, mutations in CHIP cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16), a fatal neurodegenerative disease characterized by truncal and limb ataxia that results in gait instability. Here, we systematically analyzed CHIP mutations that cause SCAR16 and found that most SCAR16 mutations destabilize CHIP. This destabilization caused mutation-specific defects in CHIP activity, including increased formation of soluble oligomers, decreased interactions with chaperones, diminished substrate ubiquitination, and reduced steady-state levels in cells. Consistent with decreased CHIP stability promoting its dysfunction in SCAR16, most mutant proteins recovered activity when the assays were performed below the mutants' melting temperature. Together, our results have uncovered the molecular basis of genetic defects in CHIP function that cause SCAR16. Our insights suggest that compounds that improve the thermostability of genetic CHIP variants may be beneficial for treating patients with SCAR16.
Keywords: molecular chaperone; neurodegenerative disease; protein misfolding; proteostasis; ubiquitin ligase.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


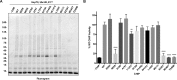
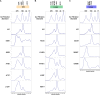
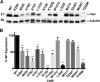

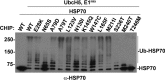
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

