Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems
- PMID: 29317671
- PMCID: PMC5760519
- DOI: 10.1038/s41598-017-18447-y
Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems
Abstract
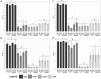
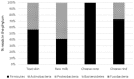
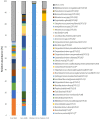
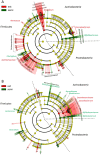
The objectives of this study were to explore bacterial community assembly from cow teat skin to raw milk cheeses and to evaluate the role of farming systems on this assembly using 16S rRNA gene high-throughput sequencing. The two grazing systems studied (extensive vs. semi-extensive) had a greater effect on the microbiota of cow teat skin than on that of raw milks and cheeses. On teat skin, the relative abundance of several taxa at different taxonomic levels (Coriobacteriia, Bifidobacteriales, Corynebacteriales, Lachnospiraceae, Atopobium, and Clostridium) varied depending on the grazing system and the period (early or late summer). In cheese, the abundance of sub-dominant lactic acid bacteria (LAB) varied depending on the grazing system. Overall, 85% of OTUs detected in raw milks and 27% of OTUs detected in ripened cheeses were also found on cow teat skin. Several shared OTUs were assigned to taxa known to be involved in the development of cheese sensory characteristics, such as Micrococcales, Staphylococcaceae, and LAB. Our results highlight the key role of cow teat skin as a reservoir of microbial diversity for raw milk, and for the first time, that cow teat skin serves as a potential source of microorganisms found in raw-milk cheeses.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Behrendt U, Müller T, Seyfarth W. The influence of extensification in grassland management on the populations of micro-organisms in the phyllosphere of grasses. Microbiol. Res. 1997;152:75–85. doi: 10.1016/S0944-5013(97)80026-2. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

