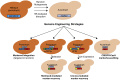
Genome and metabolic engineering in non-conventional yeasts: Current advances and applications
- PMID: 29318200
- PMCID: PMC5655347
- DOI: 10.1016/j.synbio.2017.08.002
Genome and metabolic engineering in non-conventional yeasts: Current advances and applications
Abstract
Microbial production of chemicals and proteins from biomass-derived and waste sugar streams is a rapidly growing area of research and development. While the model yeast Saccharomyces cerevisiae is an excellent host for the conversion of glucose to ethanol, production of other chemicals from alternative substrates often requires extensive strain engineering. To avoid complex and intensive engineering of S. cerevisiae, other yeasts are often selected as hosts for bioprocessing based on their natural capacity to produce a desired product: for example, the efficient production and secretion of proteins, lipids, and primary metabolites that have value as commodity chemicals. Even when using yeasts with beneficial native phenotypes, metabolic engineering to increase yield, titer, and production rate is essential. The non-conventional yeasts Kluyveromyces lactis, K. marxianus, Scheffersomyces stipitis, Yarrowia lipolytica, Hansenula polymorpha and Pichia pastoris have been developed as eukaryotic hosts because of their desirable phenotypes, including thermotolerance, assimilation of diverse carbon sources, and high protein secretion. However, advanced metabolic engineering in these yeasts has been limited. This review outlines the challenges of using non-conventional yeasts for strain and pathway engineering, and discusses the developed solutions to these problems and the resulting applications in industrial biotechnology.
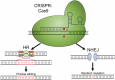
Keywords: CRISPR, Clustered regularly interspaced short palindromic repeats; CRISPR-Cas9; DSB, double strand break; HR, homologous recombination; Hansenula polymorpha; Kluyveromyces lactis; Kluyveromyces marxianus; NHEJ, nonhomologous end-joining; PAM, protospacer adjacent motif; Pichia pastoris; Scheffersomyces stipitis; TALEN, transcription activator-like effector nucleases; Yarrowia lipolytica; sgRNA, short (or single) guide RNA.
Figures
References
-
- Nielsen J., Larsson C., van Maris A., Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol. 2013;24(3):398–404. - PubMed
-
- Wagner J.M., Alper H.S. Synthetic biology and molecular genetics in non-conventional yeasts: current tools and future advances. Fungal Genet Biol. 2016;89:126–136. - PubMed
-
- Spohner S.C., Schaum V., Quitmann H., Czermak P. Kluyveromyces lactis: an emerging tool in biotechnology. J Biotechnol. 2016;222:104–116. - PubMed
-
- Varela J.A., Gethins L., Stanton C., Ross P., Morrissey J.P. Applications of Kluyveromyces marxianus in biotechnology. In: Satyanarayana T., Kunze G., editors. Yeast diversity in human welfare. Springer; Singapore: 2017. pp. 439–453.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

