Deuterostome Genomics: Lineage-Specific Protein Expansions That Enabled Chordate Muscle Evolution
- PMID: 29319812
- PMCID: PMC5888912
- DOI: 10.1093/molbev/msy002
Deuterostome Genomics: Lineage-Specific Protein Expansions That Enabled Chordate Muscle Evolution
Erratum in
-
Molecular Biology and Evolution, Volume 35, Issue 4.Mol Biol Evol. 2018 Jul 1;35(7):1821. doi: 10.1093/molbev/msy057. Mol Biol Evol. 2018. PMID: 29659984 Free PMC article. No abstract available.
Abstract
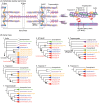
Fish-like larvae were foundational to the chordate body plan, given the basal placement of free-living lancelets. That body plan probably made it possible for chordate ancestors to swim by beating a tail formed of notochord and bilateral paraxial muscles. In order to investigate the molecular genetic basis of the origin and evolution of paraxial muscle, we deduced the evolutionary histories of 16 contractile protein genes from paraxial muscle, based on genomic data from all five deuterostome lineages, using a newly developed orthology identification pipeline and a species tree. As a result, we found that more than twice as many orthologs of paraxial muscle genes are present in chordates, as in nonchordate deuterostomes (ambulacrarians). Orthologs of paraxial-type actin and troponin C genes are absent in ambulacrarians and most paraxial muscle protein isoforms diversified via gene duplications that occurred in each chordate lineage. Analyses of genes with known expression sites indicated that some isoforms were reutilized in specific muscles of nonvertebrate chordates via gene duplications. As orthologs of most paraxial muscle genes were present in ambulacrarians, in addition to expression patterns of related genes and functions of the two protein isoforms, regulatory mechanisms of muscle genes should also be considered in future studies of the origin of paraxial muscle.
Figures


References
-
- Araki I, Saiga H, Makabe KW, Satoh N.. 1994. Expression of Amd1, a gene for a Myod1-related factor in the ascidian Halocynthia roretzi. Roux Arch Dev Biol. 2036:320–327. - PubMed
-
- Benito J, Pardos F.. 1997. Hemichordata In: Harrison FW, Ruppert EE, editors. Microscopic anatomy of invertebrates. New York: Wiley-Liss Inc; p. 15–101.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

