Discovery of Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT)
- PMID: 29320176
- PMCID: PMC5823789
- DOI: 10.1021/acs.jmedchem.7b01422
Discovery of Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT)
Erratum in
-
Correction to Discovery of Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT).J Med Chem. 2018 Jul 12;61(13):5771-5772. doi: 10.1021/acs.jmedchem.8b00849. Epub 2018 Jun 27. J Med Chem. 2018. PMID: 29947518 No abstract available.
Abstract
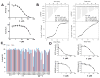
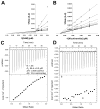
Nicotinamide N-methyltransferase (NNMT) catalyzes the N-methylation of pyridine-containing compounds using the cofactor S-5'-adenosyl-l-methionine (SAM) as the methyl group donor. Through the regulation of the levels of its substrates, cofactor, and products, NNMT plays an important role in physiology and pathophysiology. Overexpression of NNMT has been implicated in various human diseases. Potent and selective small-molecule NNMT inhibitors are valuable chemical tools for testing biological and therapeutic hypotheses. However, very few NNMT inhibitors have been reported. Here, we describe the discovery of a bisubstrate NNMT inhibitor MS2734 (6) and characterization of this inhibitor in biochemical, biophysical, kinetic, and structural studies. Importantly, we obtained the first crystal structure of human NNMT in complex with a small-molecule inhibitor. The structure of the NNMT-6 complex has unambiguously demonstrated that 6 occupied both substrate and cofactor binding sites. The findings paved the way for developing more potent and selective NNMT inhibitors in the future.
Conflict of interest statement
The authors declare the following competing financial interest(s): J.F. is an employee and shareholder of Accutar Biotechnology. J.J. is a consultant of Accutar Biotechnology.
Figures
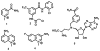
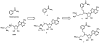



References
-
- Alston TA, Abeles RH. Substrate specificity of nicotinamide methyltransferase isolated from porcine liver. Arch Biochem Biophys. 1988;260:601–608. - PubMed
-
- van Haren MJ, Sastre Torano J, Sartini D, Emanuelli M, Parsons RB, Martin NI. A rapid and efficient assay for the characterization of substrates and inhibitors of nicotinamide N-methyltransferase. Biochemistry. 2016;55:5307–5315. - PubMed
-
- Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J Biol Chem. 1994;269:14835–14840. - PubMed
-
- Riederer M, Erwa W, Zimmermann R, Frank S, Zechner R. Adipose tissue as a source of nicotinamide N-methyltransferase and homocysteine. Atherosclerosis. 2009;204:412–417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

