Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers
- PMID: 29320577
- PMCID: PMC5761873
- DOI: 10.1371/journal.pone.0190485
Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers
Abstract
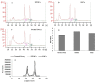
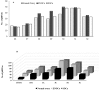
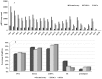
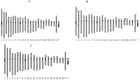
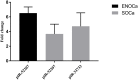
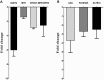
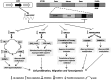
PIWI-interacting (piRNAs), ~23-36 nucleotide-long small non-coding RNAs (sncRNAs), earlier believed to be germline-specific, have now been identified in somatic cells, including cancer cells. These sncRNAs impact critical biological processes by fine-tuning gene expression at post-transcriptional and epigenetic levels. The expression of piRNAs in ovarian cancer, the most lethal gynecologic cancer is largely uncharted. In this study, we investigated the expression of PIWILs by qRT-PCR and western blotting and then identified piRNA transcriptomes in tissues of normal ovary and two most prevalent epithelial ovarian cancer subtypes, serous and endometrioid by small RNA sequencing. We detected 219, 256 and 234 piRNAs in normal ovary, endometrioid and serous ovarian cancer samples respectively. We observed piRNAs are encoded from various genomic regions, among which introns harbor the majority of them. Surprisingly, piRNAs originated from different genomic contexts showed the varied level of conservations across vertebrates. The functional analysis of predicted targets of differentially expressed piRNAs revealed these could modulate key processes and pathways involved in ovarian oncogenesis. Our study provides the first comprehensive piRNA landscape in these samples and a useful resource for further functional studies to decipher new mechanistic views of piRNA-mediated gene regulatory networks affecting ovarian oncogenesis. The RNA-seq data is submitted to GEO database (GSE83794).
Conflict of interest statement
Figures
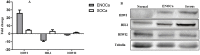



References
-
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–707. doi: 10.1158/0008-5472.CAN-07-1936 . - DOI - PubMed
-
- Devouassoux-Shisheboran M, Genestie C. Pathobiology of ovarian carcinomas. Chin J Cancer. 2015;34(1):50–5. doi: 10.5732/cjc.014.10273 - DOI - PMC - PubMed
-
- Sung PL, Chang YH, Chao KC, Chuang CM, Task Force on Systematic R, Meta-analysis of Ovarian C. Global distribution pattern of histological subtypes of epithelial ovarian cancer: a database analysis and systematic review. Gynecol Oncol. 2014;133(2):147–54. doi: 10.1016/j.ygyno.2014.02.016 . - DOI - PubMed
-
- Cunningham JM, Cicek MS, Larson NB, Davila J, Wang C, Larson MC, et al. Clinical characteristics of ovarian cancer classified by BRCA1, BRCA2, and RAD51C status. Scientific reports. 2014;4:4026 doi: 10.1038/srep04026 - DOI - PMC - PubMed
-
- Mosig RA, Lin L, Senturk E, Shah H, Huang F, Schlosshauer P, et al. Application of RNA-Seq transcriptome analysis: CD151 is an Invasion/Migration target in all stages of epithelial ovarian cancer. Journal of ovarian research. 2012;5:4 doi: 10.1186/1757-2215-5-4 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

