Variation in Activity State, Axonal Projection, and Position Define the Transcriptional Identity of Individual Neocortical Projection Neurons
- PMID: 29320739
- PMCID: PMC5863578
- DOI: 10.1016/j.celrep.2017.12.046
Variation in Activity State, Axonal Projection, and Position Define the Transcriptional Identity of Individual Neocortical Projection Neurons
Abstract
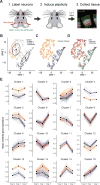
Single-cell RNA sequencing has generated catalogs of transcriptionally defined neuronal subtypes of the brain. However, the cellular processes that contribute to neuronal subtype specification and transcriptional heterogeneity remain unclear. By comparing the gene expression profiles of single layer 6 corticothalamic neurons in somatosensory cortex, we show that transcriptional subtypes primarily reflect axonal projection pattern, laminar position within the cortex, and neuronal activity state. Pseudotemporal ordering of 1,023 cellular responses to sensory manipulation demonstrates that changes in expression of activity-induced genes both reinforced cell-type identity and contributed to increased transcriptional heterogeneity within each cell type. This is due to cell-type biased choices of transcriptional states following manipulation of neuronal activity. These results reveal that axonal projection pattern, laminar position, and activity state define significant axes of variation that contribute both to the transcriptional identity of individual neurons and to the transcriptional heterogeneity within each neuronal subtype.
Keywords: activity-dependent plasticity; barrel cortex; corticothalamic neurons; neocortex; neuronal identity; single-cell RNA sequencing; somatosensory cortex; transcriptional variation.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


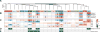

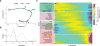

References
-
- Bourassa J, Deschênes M. Corticothalamic projections from the primary visual cortex in rats: a single fiber study using biocytin as an anterograde tracer. Neuroscience. 1995;66:253–263. - PubMed
-
- Bourassa J, Pinault D, Deschênes M. Corticothalamic projections from the cortical barrel field to the somatosensory thalamus in rats: a single-fibre study using biocytin as an anterograde tracer. Eur J Neurosci. 1995;7:19–30. - PubMed
-
- Brennecke P, Anders S, Kim JK, Ko1odziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, Heisler MG. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

