Revealing the protein propionylation activity of the histone acetyltransferase MOF (males absent on the first)
- PMID: 29321206
- PMCID: PMC5836141
- DOI: 10.1074/jbc.RA117.000529
Revealing the protein propionylation activity of the histone acetyltransferase MOF (males absent on the first)
Abstract
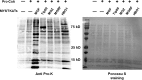
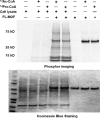
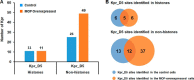
Short-chain acylation of lysine residues has recently emerged as a group of reversible posttranslational modifications in mammalian cells. The diversity of acylation further broadens the landscape and complexity of the proteome. Identification of regulatory enzymes and effector proteins for lysine acylation is critical to understand functions of these novel modifications at the molecular level. Here, we report that the MYST family of lysine acetyltransferases (KATs) possesses strong propionyltransferase activity both in vitro and in cellulo Particularly, the propionyltransferase activity of MOF, MOZ, and HBO1 is as strong as their acetyltransferase activity. Overexpression of MOF in human embryonic kidney 293T cells induced significantly increased propionylation in multiple histone and non-histone proteins, which shows that the function of MOF goes far beyond its canonical histone H4 lysine 16 acetylation. We also resolved the X-ray co-crystal structure of MOF bound with propionyl-coenzyme A, which provides a direct structural basis for the propionyltransferase activity of the MYST KATs. Our data together define a novel function for the MYST KATs as lysine propionyltransferases and suggest much broader physiological impacts for this family of enzymes.
Keywords: Males absent on the first (MOF); acetyltransferase; crystal structure; lysine propionylation; post-translational modification (PTM); protein acylation; proteomics.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




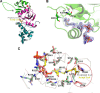

References
-
- Sabari B. R., Tang Z., Huang H., Yong-Gonzalez V., Molina H., Kong H. E., Dai L., Shimada M., Cross J. R., Zhao Y., Roeder R. G., and Allis C. D. (2015) Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 10.1016/j.molcel.2015.02.029 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

