Nonsensing residues in S3-S4 linker's C terminus affect the voltage sensor set point in K+ channels
- PMID: 29321262
- PMCID: PMC5806678
- DOI: 10.1085/jgp.201711882
Nonsensing residues in S3-S4 linker's C terminus affect the voltage sensor set point in K+ channels
Abstract
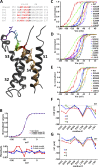
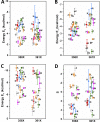
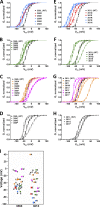
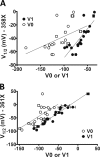
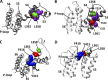
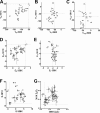
Voltage sensitivity in ion channels is a function of highly conserved arginine residues in their voltage-sensing domains (VSDs), but this conservation does not explain the diversity in voltage dependence among different K+ channels. Here we study the non-voltage-sensing residues 353 to 361 in Shaker K+ channels and find that residues 358 and 361 strongly modulate the voltage dependence of the channel. We mutate these two residues into all possible remaining amino acids (AAs) and obtain Q-V and G-V curves. We introduced the nonconducting W434F mutation to record sensing currents in all mutants except L361R, which requires K+ depletion because it is affected by W434F. By fitting Q-Vs with a sequential three-state model for two voltage dependence-related parameters (V0, the voltage-dependent transition from the resting to intermediate state and V1, from the latter to the active state) and G-Vs with a two-state model for the voltage dependence of the pore domain parameter (V1/2), Spearman's coefficients denoting variable relationships with hydrophobicity, available area, length, width, and volume of the AAs in 358 and 361 positions could be calculated. We find that mutations in residue 358 shift Q-Vs and G-Vs along the voltage axis by affecting V0, V1, and V1/2 according to the hydrophobicity of the AA. Mutations in residue 361 also shift both curves, but V0 is affected by the hydrophobicity of the AA in position 361, whereas V1 and V1/2 are affected by size-related AA indices. Small-to-tiny AAs have opposite effects on V1 and V1/2 in position 358 compared with 361. We hypothesize possible coordination points in the protein that residues 358 and 361 would temporarily and differently interact with in an intermediate state of VSD activation. Our data contribute to the accumulating knowledge of voltage-dependent ion channel activation by adding functional information about the effects of so-called non-voltage-sensing residues on VSD dynamics.
© 2018 Carvalho-de-Souza and Bezanilla.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

