HLA Class I Downregulation by HIV-1 Variants from Subtype C Transmission Pairs
- PMID: 29321314
- PMCID: PMC5972908
- DOI: 10.1128/JVI.01633-17
HLA Class I Downregulation by HIV-1 Variants from Subtype C Transmission Pairs
Abstract
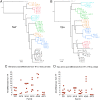
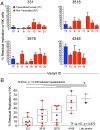
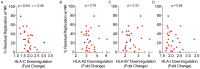
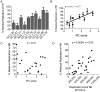
HIV-1 downregulates human leukocyte antigen A (HLA-A) and HLA-B from the surface of infected cells primarily to evade CD8 T cell recognition. HLA-C was thought to remain on the cell surface and bind inhibitory killer immunoglobulin-like receptors, preventing natural killer (NK) cell-mediated suppression. However, a recent study found HIV-1 primary viruses have the capacity to downregulate HLA-C. The goal of this study was to assess the heterogeneity of HLA-A, HLA-B, and HLA-C downregulation among full-length primary viruses from six chronically infected and six newly infected individuals from transmission pairs and to determine whether transmitted/founder variants exhibit common HLA class I downregulation characteristics. We measured HLA-A, HLA-B, HLA-C, and total HLA class I downregulation by flow cytometry of primary CD4 T cells infected with 40 infectious molecular clones. Primary viruses mediated a range of HLA class I downregulation capacities (1.3- to 6.1-fold) which could differ significantly between transmission pairs. Downregulation of HLA-C surface expression on infected cells correlated with susceptibility to in vitro NK cell suppression of virus release. Despite this, transmitted/founder variants did not share a downregulation signature and instead were more similar to the quasispecies of matched donor partners. These data indicate that a range of viral abilities to downregulate HLA-A, HLA-B, and HLA-C exist within and between individuals that can have functional consequences on immune recognition.IMPORTANCE Subtype C HIV-1 is the predominant subtype involved in heterosexual transmission in sub-Saharan Africa. Authentic subtype C viruses that contain natural sequence variations throughout the genome often are not used in experimental systems due to technical constraints and sample availability. In this study, authentic full-length subtype C viruses, including transmitted/founder viruses, were examined for the ability to disrupt surface expression of HLA class I molecules, which are central to both adaptive and innate immune responses to viral infections. We found that the HLA class I downregulation capacity of primary viruses varied, and HLA-C downregulation capacity impacted viral suppression by natural killer cells. Transmitted viruses were not distinct in the capacity for HLA class I downregulation or natural killer cell evasion. These results enrich our understanding of the phenotypic variation existing among natural HIV-1 viruses and how that might impact the ability of the immune system to recognize infected cells in acute and chronic infection.
Keywords: HIV-1; HLA; NK cells; Nef; Vpu; human immunodeficiency virus; human leukocyte antigen class I; natural killer cells; quasispecies; transmitted/founder virus.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
Resistance of Major Histocompatibility Complex Class B (MHC-B) to Nef-Mediated Downregulation Relative to that of MHC-A Is Conserved among Primate Lentiviruses and Influences Antiviral T Cell Responses in HIV-1-Infected Individuals.J Virol. 2017 Dec 14;92(1):e01409-17. doi: 10.1128/JVI.01409-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046444 Free PMC article.
-
HIV-1 Vpu Mediates HLA-C Downregulation.Cell Host Microbe. 2016 May 11;19(5):686-95. doi: 10.1016/j.chom.2016.04.005. Cell Host Microbe. 2016. PMID: 27173934 Free PMC article.
-
Convergent Evolution of HLA-C Downmodulation in HIV-1 and HIV-2.mBio. 2020 Jul 14;11(4):e00782-20. doi: 10.1128/mBio.00782-20. mBio. 2020. PMID: 32665270 Free PMC article.
-
Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins.Cytokine Growth Factor Rev. 2018 Apr;40:3-12. doi: 10.1016/j.cytogfr.2018.02.005. Epub 2018 Feb 23. Cytokine Growth Factor Rev. 2018. PMID: 29526437 Review.
-
HIV-1 accessory proteins--ensuring viral survival in a hostile environment.Cell Host Microbe. 2008 Jun 12;3(6):388-98. doi: 10.1016/j.chom.2008.04.008. Cell Host Microbe. 2008. PMID: 18541215 Review.
Cited by
-
Primary HIV-1 Strains Use Nef To Downmodulate HLA-E Surface Expression.J Virol. 2019 Sep 30;93(20):e00719-19. doi: 10.1128/JVI.00719-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31375574 Free PMC article.
-
Functional variability of Nef in antagonizing SERINC5 during acute to chronic HIV-1 infection.AIDS. 2025 Mar 1;39(3):229-240. doi: 10.1097/QAD.0000000000004079. Epub 2024 Dec 4. AIDS. 2025. PMID: 39612239 Free PMC article.
-
Ex vivo rectal explant model reveals potential opposing roles of Natural Killer cells and Marginal Zone-like B cells in HIV-1 infection.Sci Rep. 2020 Nov 19;10(1):20154. doi: 10.1038/s41598-020-76976-5. Sci Rep. 2020. PMID: 33214610 Free PMC article.
-
The Education of NK Cells Determines Their Responsiveness to Autologous HIV-Infected CD4 T Cells.J Virol. 2019 Nov 13;93(23):e01185-19. doi: 10.1128/JVI.01185-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31511383 Free PMC article.
-
HLA-C downregulation by HIV-1 adapts to host HLA genotype.PLoS Pathog. 2018 Sep 4;14(9):e1007257. doi: 10.1371/journal.ppat.1007257. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30180214 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

