Glycosyl-Phosphatidylinositol-Anchored Anti-HIV Env Single-Chain Variable Fragments Interfere with HIV-1 Env Processing and Viral Infectivity
- PMID: 29321330
- PMCID: PMC5972903
- DOI: 10.1128/JVI.02080-17
Glycosyl-Phosphatidylinositol-Anchored Anti-HIV Env Single-Chain Variable Fragments Interfere with HIV-1 Env Processing and Viral Infectivity
Abstract
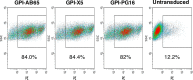
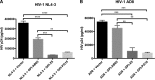
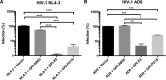
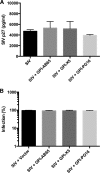
In previous studies, we demonstrated that single-chain variable fragments (scFvs) from anti-human immunodeficiency virus (HIV) Env monoclonal antibodies act as entry inhibitors when tethered to the surface of target cells by a glycosyl-phosphatidylinositol (GPI) anchor. Interestingly, even if a virus escapes inhibition at entry, its replication is ultimately controlled. We hypothesized that in addition to functioning as entry inhibitors, anti-HIV GPI-scFvs may also interact with Env in an infected cell, thereby interfering with the infectivity of newly produced virions. Here, we show that expression of the anti-HIV Env GPI-scFvs in virus-producing cells reduced the release of HIV from cells 5- to 22-fold, and infectivity of the virions that were released was inhibited by 74% to 99%. Additionally, anti-HIV Env GPI-scFv X5 inhibited virion production and infectivity after latency reactivation and blocked transmitter/founder virus production and infectivity in primary CD4+ T cells. In contrast, simian immunodeficiency virus (SIV) production and infectivity were not affected by the anti-HIV Env GPI-scFvs. Loss of infectivity of HIV was associated with a reduction in the amount of virion-associated Env gp120. Interestingly, an analysis of Env expression in cell lysates demonstrated that the anti-Env GPI-scFvs interfered with processing of Env gp160 precursors in cells. These data indicate that GPI-scFvs can inhibit Env processing and function, thereby restricting production and infectivity of newly synthesized HIV. Anti-Env GPI-scFvs therefore appear to be unique anti-HIV molecules as they derive their potent inhibitory activity by interfering with both early (receptor binding/entry) and late (Env processing and incorporation into virions) stages of the HIV life cycle.IMPORTANCE The restoration of immune function and persistence of CD4+ T cells in HIV-1-infected individuals without antiretroviral therapy requires a way to increase resistance of CD4+ T cells to infection by both R5- and X4-tropic HIV-1. Previously, we reported that anchoring anti-HIV-1 single-chain variable fragments (scFvs) via glycosyl-phosphatidylinositol (GPI) to the surface of permissive cells conferred a high level of resistance to HIV-1 variants at the level of entry. Here, we report that anti-HIV GPI-scFvs also derive their potent antiviral activity in part by blocking HIV production and Env processing, which consequently inhibits viral infectivity even in primary infection models. Thus, we conclude that GPI-anchored anti-HIV scFvs derive their potent blocking activity of HIV replication by interfering with successive stages of the viral life cycle. They may be effectively used in genetic intervention of HIV-1 infection.
Keywords: antiviral agents; entry inhibitor; envelope protein; human immunodeficiency virus; infectivity; neutralizing antibodies; scFv.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
Engineered CD4 T cells expressing a membrane anchored viral inhibitor restrict HIV-1 through cis and trans mechanisms.Front Immunol. 2023 Sep 14;14:1167965. doi: 10.3389/fimmu.2023.1167965. eCollection 2023. Front Immunol. 2023. PMID: 37781368 Free PMC article.
-
Glycosylphosphatidylinositol-Anchored Anti-HIV scFv Efficiently Protects CD4 T Cells from HIV-1 Infection and Deletion in hu-PBL Mice.J Virol. 2017 Jan 18;91(3):e01389-16. doi: 10.1128/JVI.01389-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881659 Free PMC article.
-
GPI-anchored single chain Fv--an effective way to capture transiently-exposed neutralization epitopes on HIV-1 envelope spike.Retrovirology. 2010 Oct 6;7:79. doi: 10.1186/1742-4690-7-79. Retrovirology. 2010. PMID: 20923574 Free PMC article.
-
HIV-1 diversity in the envelope glycoproteins: implications for viral entry inhibition.Viruses. 2013 Feb 6;5(2):595-604. doi: 10.3390/v5020595. Viruses. 2013. PMID: 23389465 Free PMC article. Review.
-
Evolution of Host Target Cell Specificity During HIV-1 Infection.Curr HIV Res. 2018;16(1):13-20. doi: 10.2174/1570162X16666171222105721. Curr HIV Res. 2018. PMID: 29268687 Review.
Cited by
-
Vpr Enhances HIV-1 Env Processing and Virion Infectivity in Macrophages by Modulating TET2-Dependent IFITM3 Expression.mBio. 2019 Aug 20;10(4):e01344-19. doi: 10.1128/mBio.01344-19. mBio. 2019. PMID: 31431548 Free PMC article.
-
HIV-1 subtype F integrase polymorphisms external to the catalytic core domain contribute to severe loss of replication capacity in context of the integrase inhibitor resistance mutation Q148H.J Antimicrob Chemother. 2022 Sep 30;77(10):2793-2802. doi: 10.1093/jac/dkac238. J Antimicrob Chemother. 2022. PMID: 35897124 Free PMC article.
-
Generation of HIV-resistant cells with a single-domain antibody: implications for HIV-1 gene therapy.Cell Mol Immunol. 2021 Mar;18(3):660-674. doi: 10.1038/s41423-020-00627-y. Epub 2021 Jan 18. Cell Mol Immunol. 2021. PMID: 33462383 Free PMC article.
-
Engineered CD4 T cells expressing a membrane anchored viral inhibitor restrict HIV-1 through cis and trans mechanisms.Front Immunol. 2023 Sep 14;14:1167965. doi: 10.3389/fimmu.2023.1167965. eCollection 2023. Front Immunol. 2023. PMID: 37781368 Free PMC article.
-
In vivo Serial Passaging of Human-Simian Immunodeficiency Virus Clones Identifies Characteristics for Persistent Viral Replication.Front Microbiol. 2021 Nov 18;12:779460. doi: 10.3389/fmicb.2021.779460. eCollection 2021. Front Microbiol. 2021. PMID: 34867922 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

