Blimp-1/PRDM1 is a critical regulator of Type III Interferon responses in mammary epithelial cells
- PMID: 29321612
- PMCID: PMC5762727
- DOI: 10.1038/s41598-017-18652-9
Blimp-1/PRDM1 is a critical regulator of Type III Interferon responses in mammary epithelial cells
Abstract
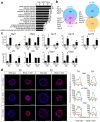
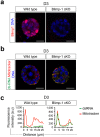
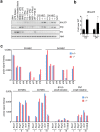
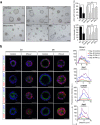
The transcriptional repressor Blimp-1 originally cloned as a silencer of type I interferon (IFN)-β gene expression controls cell fate decisions in multiple tissue contexts. Conditional inactivation in the mammary gland was recently shown to disrupt epithelial cell architecture. Here we report that Blimp-1 regulates expression of viral defense, IFN signaling and MHC class I pathways, and directly targets the transcriptional activator Stat1. Blimp-1 functional loss in 3D cultures of mammary epithelial cells (MECs) results in accumulation of dsRNA and expression of type III IFN-λ. Cultures treated with IFN lambda similarly display defective lumen formation. These results demonstrate that type III IFN-λ profoundly influences the behavior of MECs and identify Blimp-1 as a critical regulator of IFN signaling cascades.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

