Identification of Novel Signal Transduction, Immune Function, and Oxidative Stress Genes and Pathways by Topiramate for Treatment of Methamphetamine Dependence Based on Secondary Outcomes
- PMID: 29321746
- PMCID: PMC5733474
- DOI: 10.3389/fpsyt.2017.00271
Identification of Novel Signal Transduction, Immune Function, and Oxidative Stress Genes and Pathways by Topiramate for Treatment of Methamphetamine Dependence Based on Secondary Outcomes
Abstract
Background: Topiramate (TPM) is suggested to be a promising medication for treatment of methamphetamine (METH) dependence, but the molecular basis remains to be elucidated.
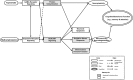
Methods: Among 140 METH-dependent participants randomly assigned to receive either TPM (N = 69) or placebo (N = 71) in a previously conducted randomized controlled trial, 50 TPM- and 49 placebo-treated participants had a total 212 RNA samples available at baseline, week 8, and week 12 time points. Following our primary analysis of gene expression data, we reanalyzed the microarray expression data based on a latent class analysis of binary secondary outcomes during weeks 1-12 that provided a classification of 21 responders and 31 non-responders with consistent responses at both time points.
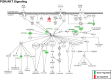
Results: Based on secondary outcomes, 1,381, 576, 905, and 711 differentially expressed genes at nominal P values < 0.05 were identified in responders versus non-responders for week 8 TPM, week 8 placebo, week 12 TPM, and week 12 placebo groups, respectively. Among 1,381 genes identified in week 8 TPM responders, 359 genes were identified in both week 8 and week 12 TPM groups, of which 300 genes were exclusively detected in TPM responders. Of them, 32 genes had nominal P values < 5 × 10-3 at either week 8 or week 12 and false discovery rates < 0.15 at both time points with consistent directions of gene expression changes, which include GABARAPL1, GPR155, and IL15RA in GABA receptor signaling that represent direct targets for TPM. Analyses of these 300 genes revealed 7 enriched pathways belonging to neuronal function/synaptic plasticity, signal transduction, inflammation/immune function, and oxidative stress response categories. No pathways were enriched for 72 genes exclusively detected in both week 8 and week 12 placebo groups.
Conclusion: This secondary analysis study of gene expression data from a TPM clinical trial not only yielded consistent results with those of primary analysis but also identified additional new genes and pathways on TPM response to METH addiction.
Keywords: clinical trial; gene expression profiling; methamphetamine dependence; microarray analysis; topiramate.
Figures


References
-
- Cho AK, Kumagai Y. Metabolism of amphetamine and other arylisopropylamines. In: Cho AK, Segal DS, editors. Amphetamine and Its Analogs: Psychopharmacology, Toxicology, and Abuse. San Diego: Academic Press; (1994). p. 43–77.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

