Development of an in vitro pre-mRNA splicing assay using plant nuclear extract
- PMID: 29321806
- PMCID: PMC5757305
- DOI: 10.1186/s13007-017-0271-6
Development of an in vitro pre-mRNA splicing assay using plant nuclear extract
Abstract
Background: Pre-mRNA splicing is an essential post-transcriptional process in all eukaryotes. In vitro splicing systems using nuclear or cytoplasmic extracts from mammalian cells, yeast, and Drosophila have provided a wealth of mechanistic insights into assembly and composition of the spliceosome, splicing regulatory proteins and mechanisms of pre-mRNA splicing in non-plant systems. The lack of an in vitro splicing system prepared from plant cells has been a major limitation in splicing research in plants.
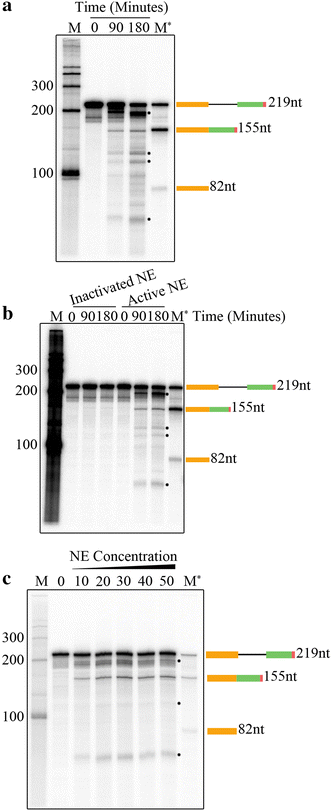
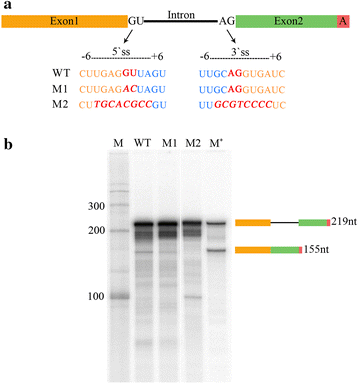
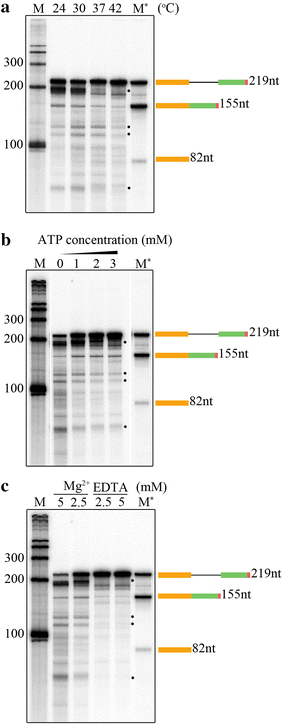
Results: Here we report an in vitro splicing assay system using plant nuclear extract. Several lines of evidence indicate that nuclear extract derived from Arabidopsis seedlings can convert pre-mRNA substrate (LHCB3) into a spliced product. These include: (1) generation of an RNA product that corresponds to the size of expected mRNA, (2) a junction-mapping assay using S1 nuclease revealed that the two exons are spliced together, (3) the reaction conditions are similar to those found with non-plant extracts and (4) finally mutations in conserved donor and acceptor sites abolished the production of the spliced product.
Conclusions: This first report on the plant in vitro splicing assay opens new avenues to investigate plant spliceosome assembly and composition, and splicing regulatory mechanisms specific to plants.
Keywords: Arabidopsis; In vitro splicing; Plant in vitro splicing; Pre-mRNA; Splicing.
Figures


References
-
- Hernandez N, Keller W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell. 1983;35(1):89–99. - PubMed
-
- Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36(4):993–1005. - PubMed
-
- Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, Sharp PA. Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225(4665):898–903. - PubMed
-
- Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260(27):14780–14792. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

