Population genomics and geographical parthenogenesis in Japanese harvestmen (Opiliones, Sclerosomatidae, Leiobunum)
- PMID: 29321849
- PMCID: PMC5756897
- DOI: 10.1002/ece3.3605
Population genomics and geographical parthenogenesis in Japanese harvestmen (Opiliones, Sclerosomatidae, Leiobunum)
Abstract
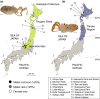
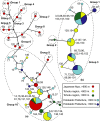
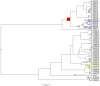
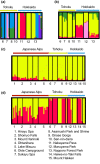
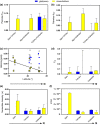
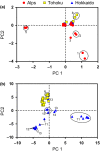
Naturally occurring population variation in reproductive mode presents an opportunity for researchers to test hypotheses regarding the evolution of sex. Asexual reproduction frequently assumes a geographical pattern, in which parthenogenesis-dominated populations are more broadly dispersed than their sexual conspecifics. We evaluate the geographical distribution of genomic signatures associated with parthenogenesis using nuclear and mitochondrial DNA sequence data from two Japanese harvestman sister taxa, Leiobunum manubriatum and Leiobunum globosum. Asexual reproduction is putatively facultative in these species, and female-biased localities are common in habitat margins. Past karyotypic and current cytometric work indicates L. globosum is entirely tetraploid, while L. manubriatum may be either diploid or tetraploid. We estimated species phylogeny, genetic differentiation, diversity, and mitonuclear discordance in females collected across the species range in order to identify range expansion toward marginal habitat, potential for hybrid origin, and persistence of asexual lineages. Our results point to northward expansion of a tetraploid ancestor of L. manubriatum and L. globosum, coupled with support for greater male gene flow in southern L. manubriatum localities. Specimens from localities in the Tohoku and Hokkaido regions were indistinct, particularly those of L. globosum, potentially due to little mitochondrial differentiation or haplotypic variation. Although L. manubriatum overlaps with L. globosum across its entire range, L. globosum was reconstructed as monophyletic with strong support using mtDNA, and marginal support with nuclear loci. Ultimately, we find evidence for continued sexual reproduction in both species and describe opportunities to clarify the rate and mechanism of parthenogenesis.
Keywords: Opiliones; geographical parthenogenesis; mitonuclear discordance; population genomics.
Figures


References
-
- Ashman, T. L. , Bachtrog, D. , Blackmon, H. , Goldberg, E. E. , Hahn, M. W. , Kirkpatrick, M. , … Vamosi, J. C. (2014). Tree of sex: A database of sexual systems. Scientific Data, 1, 140015 https://doi.org/10.1038/sdata.2014.15 - DOI - PMC - PubMed
-
- Baldo, L. , Prendini, L. , Corthals, A. , & Werren, J. H. (2007). Wolbachia are present in southern African scorpions and cluster with supergroup F. Current Microbiology, 55, 367–373. https://doi.org/10.1007/s00284-007-9009-4 - DOI - PubMed
-
- Bandelt, H. , Forster, P. , & Röhl, A. (1999). Median‐joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1), 37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036 - DOI - PubMed
-
- Barrow, L. N. , Bigelow, A. T. , Phillips, C. A. , & Lemmon, E. M. (2015). Phylogeographic inference using Bayesian model comparison across a fragmented chorus frog species complex. Molecular Ecology, 24, 4739–4758. https://doi.org/10.1111/mec.13343 - DOI - PubMed
-
- Berthelot, C. , Brunet, F. , Chalopin, D. , Juanchich, A. , Bernard, M. , Noël, B. , … Guiguen, Y. (2014). The rainbow trout genome provides novel insights into evolution after whole‐genome duplication in vertebrates. Nature Communications, 5, 3657 https://doi.org/10.1038/ncomms4657 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

