Using Genome Sequence to Enable the Design of Medicines and Chemical Probes
- PMID: 29322778
- PMCID: PMC5989578
- DOI: 10.1021/acs.chemrev.7b00504
Using Genome Sequence to Enable the Design of Medicines and Chemical Probes
Abstract
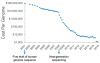
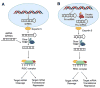
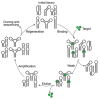
Rapid progress in genome sequencing technology has put us firmly into a postgenomic era. A key challenge in biomedical research is harnessing genome sequence to fulfill the promise of personalized medicine. This Review describes how genome sequencing has enabled the identification of disease-causing biomolecules and how these data have been converted into chemical probes of function, preclinical lead modalities, and ultimately U.S. Food and Drug Administration (FDA)-approved drugs. In particular, we focus on the use of oligonucleotide-based modalities to target disease-causing RNAs; small molecules that target DNA, RNA, or protein; the rational repurposing of known therapeutic modalities; and the advantages of pharmacogenetics. Lastly, we discuss the remaining challenges and opportunities in the direct utilization of genome sequence to enable design of medicines.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



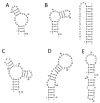

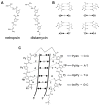
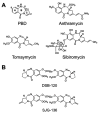
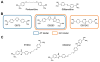
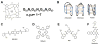


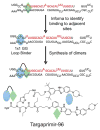


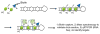
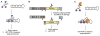

References
-
- Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A. Complete Nucleotide Sequence of Bacteriophage Ms2 Rna: Primary and Secondary Structure of the Replicase Gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

