Comparison, alignment, and synchronization of cell line information between CLO and EFO
- PMID: 29322915
- PMCID: PMC5763470
- DOI: 10.1186/s12859-017-1979-z
Comparison, alignment, and synchronization of cell line information between CLO and EFO
Abstract
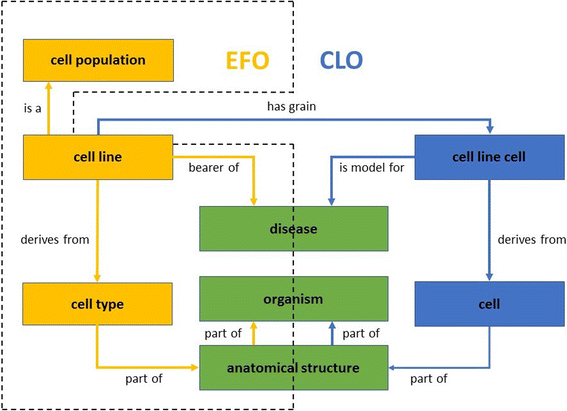
Background: The Experimental Factor Ontology (EFO) is an application ontology driven by experimental variables including cell lines to organize and describe the diverse experimental variables and data resided in the EMBL-EBI resources. The Cell Line Ontology (CLO) is an OBO community-based ontology that contains information of immortalized cell lines and relevant experimental components. EFO integrates and extends ontologies from the bio-ontology community to drive a number of practical applications. It is desirable that the community shares design patterns and therefore that EFO reuses the cell line representation from the Cell Line Ontology (CLO). There are, however, challenges to be addressed when developing a common ontology design pattern for representing cell lines in both EFO and CLO.
Results: In this study, we developed a strategy to compare and map cell line terms between EFO and CLO. We examined Cellosaurus resources for EFO-CLO cross-references. Text labels of cell lines from both ontologies were verified by biological information axiomatized in each source. The study resulted in the identification 873 EFO-CLO aligned and 344 EFO unique immortalized permanent cell lines. All of these cell lines were updated to CLO and the cell line related information was merged. A design pattern that integrates EFO and CLO was also developed.
Conclusion: Our study compared, aligned, and synchronized the cell line information between CLO and EFO. The final updated CLO will be examined as the candidate ontology to import and replace eligible EFO cell line classes thereby supporting the interoperability in the bio-ontology domain. Our mapping pipeline illustrates the use of ontology in aiding biological data standardization and integration through the biological and semantics content of cell lines.
Keywords: Cell line; Cell line ontology; Data integration; Data mapping; Experimental factor ontology.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol [Internet]. 2012;13:R5. Available from: http://genomebiology.biomedcentral.com/articles/10.1186/gb-2012-13-1-r5. - DOI - PMC - PubMed
-
- Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, et al. CLO: the cell line ontology. J Biomed Semantics [Internet]. 2014;5:37. Available from: http://jbiomedsem.biomedcentral.com/articles/10.1186/2041-1480-5-37. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

