Clonal analysis of lineage fate in native haematopoiesis
- PMID: 29323290
- PMCID: PMC5884107
- DOI: 10.1038/nature25168
Clonal analysis of lineage fate in native haematopoiesis
Abstract
Haematopoiesis, the process of mature blood and immune cell production, is functionally organized as a hierarchy, with self-renewing haematopoietic stem cells and multipotent progenitor cells sitting at the very top. Multiple models have been proposed as to what the earliest lineage choices are in these primitive haematopoietic compartments, the cellular intermediates, and the resulting lineage trees that emerge from them. Given that the bulk of studies addressing lineage outcomes have been performed in the context of haematopoietic transplantation, current models of lineage branching are more likely to represent roadmaps of lineage potential than native fate. Here we use transposon tagging to clonally trace the fates of progenitors and stem cells in unperturbed haematopoiesis. Our results describe a distinct clonal roadmap in which the megakaryocyte lineage arises largely independently of other haematopoietic fates. Our data, combined with single-cell RNA sequencing, identify a functional hierarchy of unilineage- and oligolineage-producing clones within the multipotent progenitor population. Finally, our results demonstrate that traditionally defined long-term haematopoietic stem cells are a significant source of megakaryocyte-restricted progenitors, suggesting that the megakaryocyte lineage is the predominant native fate of long-term haematopoietic stem cells. Our study provides evidence for a substantially revised roadmap for unperturbed haematopoiesis, and highlights unique properties of multipotent progenitors and haematopoietic stem cells in situ.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
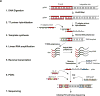






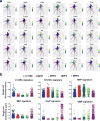






References
-
- Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. 1997;124:1929–1939. - PubMed
-
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

