Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders
- PMID: 29323667
- PMCID: PMC5912505
- DOI: 10.1038/gim.2017.246
Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders
Abstract
Purpose: Given the rapid pace of discovery in rare disease genomics, it is likely that improvements in diagnostic yield can be made by systematically reanalyzing previously generated genomic sequence data in light of new knowledge.
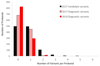
Methods: We tested this hypothesis in the United Kingdom-wide Deciphering Developmental Disorders study, where in 2014 we reported a diagnostic yield of 27% through whole-exome sequencing of 1,133 children with severe developmental disorders and their parents. We reanalyzed existing data using improved variant calling methodologies, novel variant detection algorithms, updated variant annotation, evidence-based filtering strategies, and newly discovered disease-associated genes.
Results: We are now able to diagnose an additional 182 individuals, taking our overall diagnostic yield to 454/1,133 (40%), and another 43 (4%) have a finding of uncertain clinical significance. The majority of these new diagnoses are due to novel developmental disorder-associated genes discovered since our original publication.
Conclusion: This study highlights the importance of coupling large-scale research with clinical practice, and of discussing the possibility of iterative reanalysis and recontact with patients and health professionals at an early stage. We estimate that implementing parent-offspring whole-exome sequencing as a first-line diagnostic test for developmental disorders would diagnose >50% of patients.
Keywords: diagnostic yield; exome sequencing; reanalysis; reclassification; recontact.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

