Network perturbation analysis of gene transcriptional profiles reveals protein targets and mechanism of action of drugs and influenza A viral infection
- PMID: 29325153
- PMCID: PMC5887474
- DOI: 10.1093/nar/gkx1314
Network perturbation analysis of gene transcriptional profiles reveals protein targets and mechanism of action of drugs and influenza A viral infection
Abstract
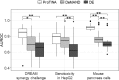
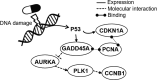
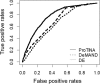
Genome-wide transcriptional profiling provides a global view of cellular state and how this state changes under different treatments (e.g. drugs) or conditions (e.g. healthy and diseased). Here, we present ProTINA (Protein Target Inference by Network Analysis), a network perturbation analysis method for inferring protein targets of compounds from gene transcriptional profiles. ProTINA uses a dynamic model of the cell-type specific protein-gene transcriptional regulation to infer network perturbations from steady state and time-series differential gene expression profiles. A candidate protein target is scored based on the gene network's dysregulation, including enhancement and attenuation of transcriptional regulatory activity of the protein on its downstream genes, caused by drug treatments. For benchmark datasets from three drug treatment studies, ProTINA was able to provide highly accurate protein target predictions and to reveal the mechanism of action of compounds with high sensitivity and specificity. Further, an application of ProTINA to gene expression profiles of influenza A viral infection led to new insights of the early events in the infection.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

