Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses
- PMID: 29325581
- PMCID: PMC5765694
- DOI: 10.1186/s40168-017-0395-y
Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses
Abstract
Background: While almost all infants are infected with respiratory syncytial virus (RSV) before the age of 2 years, only a small percentage develops severe disease. Previous studies suggest that the nasopharyngeal microbiome affects disease development. We therefore studied the effect of the nasopharyngeal microbiome on viral load and mucosal cytokine responses, two important factors influencing the pathophysiology of RSV disease. To determine the relation between (i) the microbiome of the upper respiratory tract, (ii) viral load, and (iii) host mucosal inflammation during an RSV infection, nasopharyngeal microbiota profiles of RSV infected infants (< 6 months) with different levels of disease severity and age-matched healthy controls were determined by 16S rRNA marker gene sequencing. The viral load was measured using qPCR. Nasopharyngeal CCL5, CXCL10, MMP9, IL6, and CXCL8 levels were determined with ELISA.
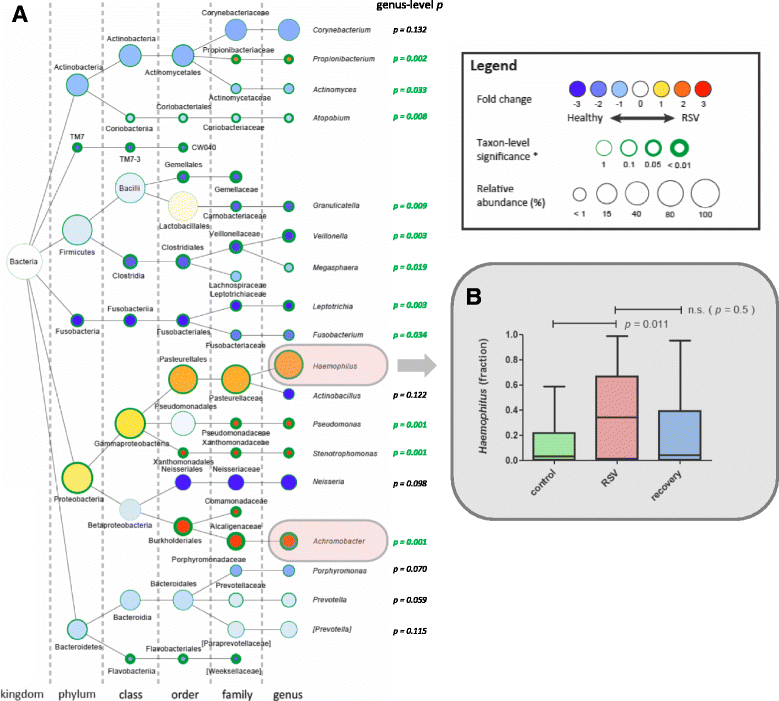
Results: Viral load in nasopharyngeal aspirates of patients associates significantly to total nasopharyngeal microbiota composition. Healthy infants (n = 21) and RSV patients (n = 54) display very distinct microbial patterns, primarily characterized by a loss in commensals like Veillonella and overrepresentation of opportunistic organisms like Haemophilus and Achromobacter in RSV-infected individuals. Furthermore, nasopharyngeal microbiota profiles are significantly different based on CXCL8 levels. CXCL8 is a chemokine that was previously found to be indicative for disease severity and for which we find Haemophilus abundance as the strongest predictor for CXCL8 levels.
Conclusions: The nasopharyngeal microbiota in young infants with RSV infection is marked by an overrepresentation of the genus Haemophilus. We present that this bacterium is associated with viral load and mucosal CXCL8 responses, both which are involved in RSV disease pathogenesis.
Keywords: Chemokine; Microbiome; Mucosal inflammation; RSV; Viral load.
Conflict of interest statement
Ethics approval and consent to participate
The study was approved by the Central Committee on Research Involving Human Subjects of the Radboud University Medical Center.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. The funding sources had no role in the design of the study, nor in collection, management, analysis, and interpretation of the data; did not partake in preparation, review, or approval of the manuscript; and had no decision in submitting the manuscript for publication.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

