Production of HIV-1 vif mRNA Is Modulated by Natural Nucleotide Variations and SLSA1 RNA Structure in SA1D2prox Genomic Region
- PMID: 29326677
- PMCID: PMC5741601
- DOI: 10.3389/fmicb.2017.02542
Production of HIV-1 vif mRNA Is Modulated by Natural Nucleotide Variations and SLSA1 RNA Structure in SA1D2prox Genomic Region
Abstract
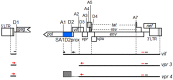
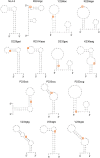
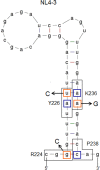
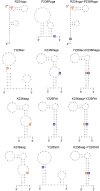
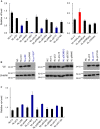
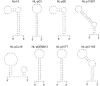
Genomic RNA of HIV-1 contains localized structures critical for viral replication. Its structural analysis has demonstrated a stem-loop structure, SLSA1, in a nearby region of HIV-1 genomic splicing acceptor 1 (SA1). We have previously shown that the expression level of vif mRNA is considerably altered by some natural single-nucleotide variations (nSNVs) clustering in SLSA1 structure. In this study, besides eleven nSNVs previously identified by us, we totally found nine new nSNVs in the SLSA1-containing sequence from SA1, splicing donor 2, and through to the start codon of Vif that significantly affect the vif mRNA level, and designated the sequence SA1D2prox (142 nucleotides for HIV-1 NL4-3). We then examined by extensive variant and mutagenesis analyses how SA1D2prox sequence and SLSA1 secondary structure are related to vif mRNA level. While the secondary structure and stability of SLSA1 was largely changed by nSNVs and artificial mutations introduced to restore the original NL4-3 form from altered ones by nSNVs, no clear association of the two SLSA1 properties with vif mRNA level was observed. In contrast, when naturally occurring SA1D2prox sequences that contain multiple nSNVs were examined, we attained significant inverse correlation between the vif level and SLSA1 stability. These results may suggest that SA1D2prox sequence adapts over time, and also that the altered SA1D2prox sequence, SLSA1 stability, and vif level are mutually related. In total, we show here that the entire SA1D2prox sequence and SLSA1 stability critically contribute to the modulation of vif mRNA level.
Keywords: HIV-1; SA1; SA1D2prox; SLSA1; nNSV; secondary RNA structure; vif mRNA.
Figures





Similar articles
-
Expression Level of HIV-1 Vif Can Be Fluctuated by Natural Nucleotide Variations in the vif-Coding and Regulatory SA1D2prox Sequences of the Proviral Genome.Front Microbiol. 2019 Nov 28;10:2758. doi: 10.3389/fmicb.2019.02758. eCollection 2019. Front Microbiol. 2019. PMID: 31849897 Free PMC article.
-
The Expression Level of HIV-1 Vif Is Optimized by Nucleotide Changes in the Genomic SA1D2prox Region during the Viral Adaptation Process.Viruses. 2021 Oct 15;13(10):2079. doi: 10.3390/v13102079. Viruses. 2021. PMID: 34696508 Free PMC article.
-
Natural Single-Nucleotide Variations in the HIV-1 Genomic SA1prox Region Can Alter Viral Replication Ability by Regulating Vif Expression Levels.J Virol. 2016 Apr 14;90(9):4563-4578. doi: 10.1128/JVI.02939-15. Print 2016 May. J Virol. 2016. PMID: 26912631 Free PMC article.
-
The role of Vif oligomerization and RNA chaperone activity in HIV-1 replication.Virus Res. 2012 Nov;169(2):361-76. doi: 10.1016/j.virusres.2012.06.018. Epub 2012 Jun 21. Virus Res. 2012. PMID: 22728817 Review.
-
The role of Vif during HIV-1 infection: interaction with novel host cellular factors.J Clin Virol. 2003 Feb;26(2):143-52. doi: 10.1016/s1386-6532(02)00113-0. J Clin Virol. 2003. PMID: 12600646 Review.
Cited by
-
Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation.Viruses. 2023 Dec 14;15(12):2424. doi: 10.3390/v15122424. Viruses. 2023. PMID: 38140666 Free PMC article.
-
Commentary: Derivation of Simian Tropic HIV-1 Infectious Clone Reveals Virus Adaptation to a New Host.Front Cell Infect Microbiol. 2020 May 15;10:235. doi: 10.3389/fcimb.2020.00235. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32500043 Free PMC article. No abstract available.
-
Expression Level of HIV-1 Vif Can Be Fluctuated by Natural Nucleotide Variations in the vif-Coding and Regulatory SA1D2prox Sequences of the Proviral Genome.Front Microbiol. 2019 Nov 28;10:2758. doi: 10.3389/fmicb.2019.02758. eCollection 2019. Front Microbiol. 2019. PMID: 31849897 Free PMC article.
-
The Expression Level of HIV-1 Vif Is Optimized by Nucleotide Changes in the Genomic SA1D2prox Region during the Viral Adaptation Process.Viruses. 2021 Oct 15;13(10):2079. doi: 10.3390/v13102079. Viruses. 2021. PMID: 34696508 Free PMC article.
-
A scale-free analysis of the HIV-1 genome demonstrates multiple conserved regions of structural and functional importance.PLoS Comput Biol. 2019 Sep 23;15(9):e1007345. doi: 10.1371/journal.pcbi.1007345. eCollection 2019 Sep. PLoS Comput Biol. 2019. PMID: 31545786 Free PMC article.
References
-
- Akari H., Fujita M., Kao S., Khan M. A., Shehu-Xhilaga M., Adachi A., et al. (2004). High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 279 12355–12362. 10.1074/jbc.M312426200 - DOI - PubMed
-
- Akari H., Uchiyama T., Fukumori T., Iida S., Koyama A. H., Adachi A. (1999). Pseudotyping human immunodeficiency virus type 1 by vesicular stomatitis virus G protein does not reduce the cell-dependent requirement of Vif for optimal infectivity: functional difference between Vif and Nef. J. Gen. Virol. 80 (Pt 11), 2945–2949. 10.1099/0022-1317-80-11-2945 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

