Accuracy of Programs for the Determination of Human Leukocyte Antigen Alleles from Next-Generation Sequencing Data
- PMID: 29326702
- PMCID: PMC5733459
- DOI: 10.3389/fimmu.2017.01815
Accuracy of Programs for the Determination of Human Leukocyte Antigen Alleles from Next-Generation Sequencing Data
Abstract
The human leukocyte antigen (HLA) genes code for proteins that play a central role in the function of the immune system by presenting peptide antigens to T cells. As HLA genes show extremely high genetic polymorphism, HLA typing at the allele level is demanding and is based on DNA sequencing. Determination of HLA alleles is warranted as HLA alleles are major genetic risk factors in autoimmune diseases and are matched in transplantation. Here, we compared the accuracy of several published HLA-typing algorithms that are based on next-generation sequencing (NGS) data. As genome sequencing is becoming increasingly common in research, we wanted to test how well HLA alleles can be deduced from genome data produced in studies with objectives other than HLA typing and in platforms not especially designed for HLA typing. The accuracies were assessed using datasets consisting of NGS data produced using an in-house sequencing platform, including the full 4 Mbp HLA segment, from 94 stem cell transplantation patients and exome sequences from 63 samples of the 1000 Genomes collection. In the patient dataset, none of the software gave perfect results for all the samples and genes when programs were used with the default settings. However, we found that ensemble prediction of the results or modifications of the settings could be used to improve accuracy. For the exome-only data, most of the algorithms did not perform very well. The results indicate that the use of these algorithms for accurate HLA allele determination is not straightforward when based on NGS data not especially targeted to the HLA typing and their accurate use requires HLA expertise.
Keywords: genetic variation; genome sequence; histocompatibility; human leukocyte antigen alleles; transplantation.
Figures
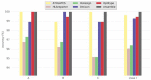
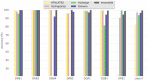
References
-
- Charron D, Petersdorf E. The HLA system in hematopoietic stem cell transplantation. 1st ed In: Socié G, Blazar B, editors. Immune Biology of Allogeneic Hematopoietic Stem Cell Transplantation. London: Elsevier; (2013). p. 19–38.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

