Chronic oxymatrine treatment induces resistance and epithelial‑mesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells
- PMID: 29328404
- PMCID: PMC5802036
- DOI: 10.3892/or.2018.6204
Chronic oxymatrine treatment induces resistance and epithelial‑mesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells
Retraction in
-
[Retracted] Chronic oxymatrine treatment induces resistance and epithelial‑mesenchymal transition through targeting the long non‑coding RNA MALAT1 in colorectal cancer cells.Oncol Rep. 2024 Jun;51(6):73. doi: 10.3892/or.2024.8732. Epub 2024 Apr 12. Oncol Rep. 2024. PMID: 38606514 Free PMC article.
Abstract
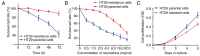
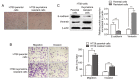
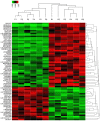
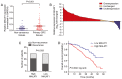
A major reason for colorectal cancer (CRC) chemoresistance is the enhanced migration and invasion of cancer cells, such as the cell acquisition of epithelial-mesenchymal transition (EMT). Long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been considered as a pro-oncogene in multiple cancers. However, the precise functional mechanism of lncRNA MALAT1 in chemoresistance and EMT is not well known. In the present study, we focused on the effect of oxymatrine on CRC cells and further investigated the role of MALAT1 in oxymatrine-induced resistance and EMT process. The human CRC cell line HT29 was exposed to increasing doses of oxymatrine to establish stable cell lines resistant to oxymatrine. The established HT29 oxymatrine resistant cells showed an EMT phenotype including specific morphologic changes, enhanced migratory and invasive capacity, and downregulation of E-cadherin protein expression. Subsequently, high-throughput HiSeq sequencing and RT-qPCR showed that lncRNA MALAT1 was significantly upregulated in the oxymatrine resistant cells (P<0.01), while knockdown of MALAT1 partially reversed the EMT phenotype in HT29 resistant cells. Furthermore, oxymatrine treatment suppressed the migration and invasion ability of CRC cells, however, this effect was significantly reversed by overexpression of MALAT1. Finally, we investigated the clinical role of MALAT1 and found that high lncRNA MALAT1 expression level is associated with poor prognosis in CRC patients receiving oxymatrine treatment (P<0.01). In conclusion, we demonstrate that lncRNA MALAT1 is a stimulator for oxymatrine resistance in CRC and it may provide therapeutic and prognostic information for CRC patients.
Figures



References
-
- Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, Wang CX. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. - PubMed
-
- Alberts SR, Horvath WL, Sternfeld WC, Goldberg RM, Mahoney MR, Dakhil SR, Levitt R, Rowland K, Nair S, Sargent DJ, et al. Oxaliplatin, fluorouracil, and leucovorin for patients with unresectable liver-only metastases from colorectal cancer: A North Central Cancer Treatment Group phase II study. J Clin Oncol. 2005;23:9243–9249. doi: 10.1200/JCO.2005.07.740. - DOI - PubMed
-
- Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

