Association of extracellular dNTP utilization with a GmPAP1-like protein identified in cell wall proteomic analysis of soybean roots
- PMID: 29329437
- PMCID: PMC5853315
- DOI: 10.1093/jxb/erx441
Association of extracellular dNTP utilization with a GmPAP1-like protein identified in cell wall proteomic analysis of soybean roots
Abstract
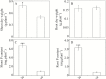
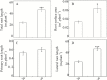
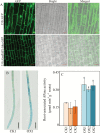
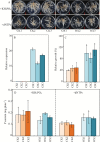
Plant root cell walls are dynamic systems that serve as the first plant compartment responsive to soil conditions, such as phosphorus (P) deficiency. To date, evidence for the regulation of root cell wall proteins (CWPs) by P deficiency remains sparse. In order to gain a better understanding of the roles played by CWPs in the roots of soybean (Glycine max) in adaptation to P deficiency, we conducted an iTRAQ (isobaric tag for relative and absolute quantitation) proteomic analysis. A total of 53 CWPs with differential accumulation in response to P deficiency were identified. Subsequent qRT-PCR analysis correlated the accumulation of 21 of the 27 up-regulated proteins, and eight of the 26 down-regulated proteins with corresponding gene expression patterns in response to P deficiency. One up-regulated CWP, purple acid phosphatase 1-like (GmPAP1-like), was functionally characterized. Phaseolus vulgaris transgenic hairy roots overexpressing GmPAP1-like displayed an increase in root-associated acid phosphatase activity. In addition, relative growth and P content were significantly enhanced in GmPAP1-like overexpressing lines compared to control lines when deoxy-ribonucleotide triphosphate (dNTP) was applied as the sole external P source. Taken together, the results suggest that the modulation of CWPs may regulate complex changes in the root system in response to P deficiency, and that the cell wall-localized GmPAP1-like protein is involved in extracellular dNTP utilization in soybean.
Keywords: Cell wall proteins; phosphorus deficiency; phosphorus utilization; proteomics; purple acid phosphatase; soybean.
Figures



References
-
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF. 2000. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. The Plant Journal 24, 457–466. - PubMed
-
- Ao JH, Fu JB, Tian J, Yan XL, Liao H. 2010. Genetic variability for root morph-architecture traits and root growth dynamics as related to phosphorus efficiency in soybean. Functional Plant Biology 37, 304–312.
-
- Asmar F, Gahoonia T, Nielsen NE. 1995. Barley genotypes differ in activity of soluble extracellular phosphatase and depletion of organic phosphorus in the rhizosphere soil. Plant and Soil 172, 117–122.
-
- Baiya S, Hua Y, Ekkhara W, Ketudat Cairns JR. 2014. Expression and enzymatic properties of rice (Oryza sativa L.) monolignol β-glucosidases. Plant Science 227, 101–109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

