Accuracy of four mononucleotide-repeat markers for the identification of DNA mismatch-repair deficiency in solid tumors
- PMID: 29329588
- PMCID: PMC5767035
- DOI: 10.1186/s12967-017-1376-4
Accuracy of four mononucleotide-repeat markers for the identification of DNA mismatch-repair deficiency in solid tumors
Abstract
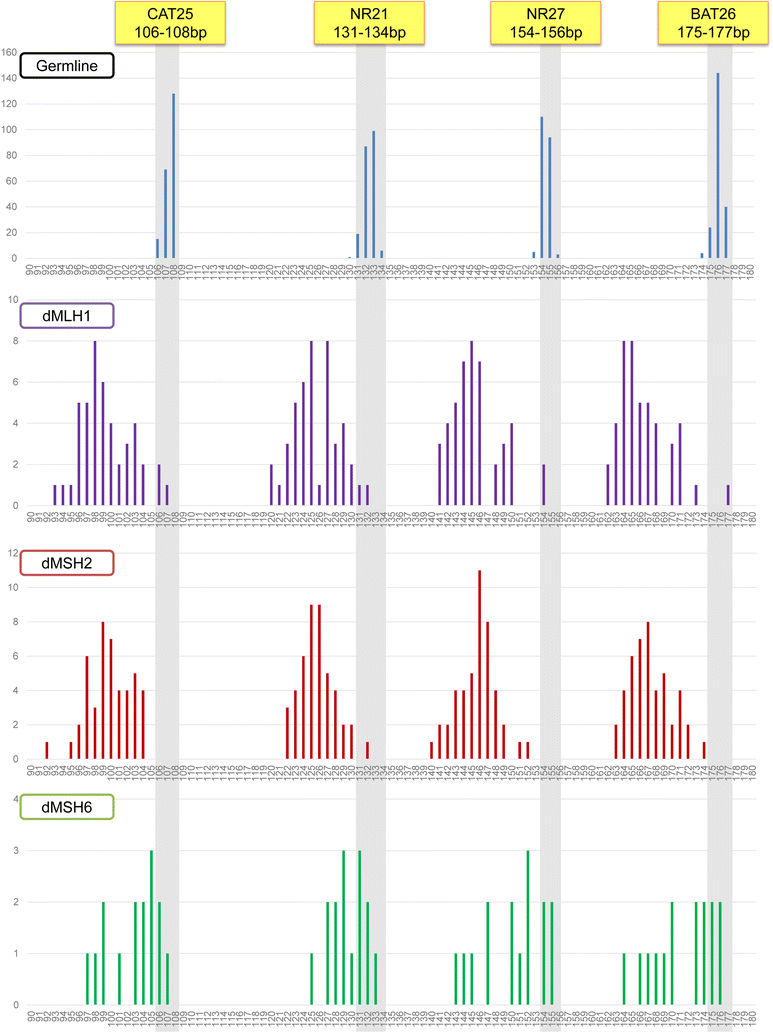
Background: To screen tumors with microsatellite instability (MSI) arising due to DNA mismatch repair deficiency (dMMR), a panel of five quasi-monomorphic mononucleotide-repeat markers amplified in a multiplex PCR (Pentaplex) are commonly used. In spite of its several strengths, the pentaplex assay is not robust at detecting the loss of MSH6-deficiency (dMSH6). In order to overcome this challenge, we designed this study to develop and optimize a panel of four quasi-monomorphic mononucleotide-repeat markers (Tetraplex) for identifying solid tumors with dMMR, especially dMSH6.
Methods: To improve the sensitivity for tumors with dMMR, we established a quasi-monomorphic variant range (QMVR) of 3-4 bp for the four Tetraplex markers. Thereafter, to confirm the accuracy of this assay, we examined 317 colorectal cancer (CRC) specimens, comprising of 105 dMMR [45 MutL homolog (MLH)1-deficient, 45 MutS protein homolog (MSH)2-deficient, and 15 MSH6-deficient tumors] and 212 MMR-proficient (pMMR) tumors as a test set. In addition, we analyzed a cohort of 138 endometrial cancers (EC) by immunohistochemistry to determine MMR protein expression and validation of our new MSI assay.
Results: Using the criteria of ≥ 1 unstable markers as MSI-positive tumor, our assay resulted in a sensitivity of 97.1% [95% confidence interval (CI) = 91.9-99.0%] for dMMR, and a specificity of 95.3% (95% CI = 91.5-97.4%) for pMMR CRC specimens. Among the 138 EC specimens, 41 were dMMR according to immunohistochemistry. Herein, our Tetraplex assay detected dMMR tumors with a sensitivity of 92.7% (95% CI = 80.6-97.5%) and a specificity of 97.9% (95% CI = 92.8-99.4%) for pMMR tumors. With respect to tumors with dMSH6, in the CRC-validation set, Tetraplex detected dMSH6 tumors with a sensitivity of 86.7% (13 of 15 dMSH6 CRCs), which was subsequently validated in the EC test set as well (sensitivity, 75.0%; 6 of 8 dMSH6 ECs).
Conclusions: Our newly optimized Tetraplex system will help offer a robust and highly sensitive assay for the identification of dMMR in solid tumors.
Keywords: Colorectal cancer; DNA mismatch repair; Endometrial cancer; Hypermutated tumors; Microsatellite instability.
Figures



References
-
- Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA72851/Division of Cancer Epidemiology and Genetics, National Cancer Institute/International
- 25860409/Japan Society for the Promotion of Science/International
- 15H03034/Japan Society for the Promotion of Science/International
- R01 CA202797/CA/NCI NIH HHS/United States
- CA202797/Division of Cancer Epidemiology and Genetics, National Cancer Institute/International
- CA184792/Division of Cancer Epidemiology and Genetics, National Cancer Institute/International
- R01 CA184792/CA/NCI NIH HHS/United States
- U01 CA187956/CA/NCI NIH HHS/United States
- R01 CA181572/CA/NCI NIH HHS/United States
- 26462016/Japan Society for the Promotion of Science/International
- R01 CA072851/CA/NCI NIH HHS/United States
- RP140784/Cancer Prevention and Research Institute of Texas/International
- CA181572/Division of Cancer Epidemiology and Genetics, National Cancer Institute (US)/International
- 20590572/Japan Society for the Promotion of Science/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

