Feline coronavirus: Insights into viral pathogenesis based on the spike protein structure and function
- PMID: 29329682
- PMCID: PMC7112122
- DOI: 10.1016/j.virol.2017.12.027
Feline coronavirus: Insights into viral pathogenesis based on the spike protein structure and function
Abstract
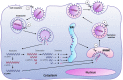
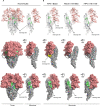
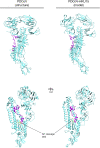
Feline coronavirus (FCoV) is an etiological agent that causes a benign enteric illness and the fatal systemic disease feline infectious peritonitis (FIP). The FCoV spike (S) protein is considered the viral regulator for binding and entry to the cell. This protein is also involved in FCoV tropism and virulence, as well as in the switch from enteric disease to FIP. This regulation is carried out by spike's major functions: receptor binding and virus-cell membrane fusion. In this review, we address important aspects in FCoV genetics, replication and pathogenesis, focusing on the role of S. To better understand this, FCoV S protein models were constructed, based on the human coronavirus NL63 (HCoV-NL63) S structure. We describe the specific structural characteristics of the FCoV S, in comparison with other coronavirus spikes. We also revise the biochemical events needed for FCoV S activation and its relation to the structural features of the protein.
Keywords: Cleavage activation; Coronavirus; Feline coronavirus; Feline infectious peritonitis; Pathogenesis; Serotype; Spike protein; Spike structure; Tropism.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

