Conformation and Trimer Association of the Transmembrane Domain of the Parainfluenza Virus Fusion Protein in Lipid Bilayers from Solid-State NMR: Insights into the Sequence Determinants of Trimer Structure and Fusion Activity
- PMID: 29330069
- PMCID: PMC5831503
- DOI: 10.1016/j.jmb.2018.01.002
Conformation and Trimer Association of the Transmembrane Domain of the Parainfluenza Virus Fusion Protein in Lipid Bilayers from Solid-State NMR: Insights into the Sequence Determinants of Trimer Structure and Fusion Activity
Abstract
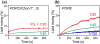
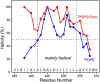
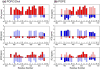
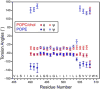
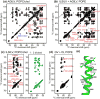
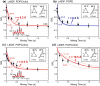
Enveloped viruses enter cells by using their fusion proteins to merge the virus lipid envelope and the cell membrane. While crystal structures of the water-soluble ectodomains of many viral fusion proteins have been determined, the structure and assembly of the C-terminal transmembrane domain (TMD) remains poorly understood. Here we use solid-state NMR to determine the backbone conformation and oligomeric structure of the TMD of the parainfluenza virus 5 fusion protein. 13C chemical shifts indicate that the central leucine-rich segment of the TMD is α-helical in POPC/cholesterol membranes and POPE membranes, while the Ile- and Val-rich termini shift to the β-strand conformation in the POPE membrane. Importantly, lipid mixing assays indicate that the TMD is more fusogenic in the POPE membrane than in the POPC/cholesterol membrane, indicating that the β-strand conformation is important for fusion by inducing membrane curvature. Incorporation of para-fluorinated Phe at three positions of the α-helical core allowed us to measure interhelical distances using 19F spin diffusion NMR. The data indicate that, at peptide:lipid molar ratios of ~1:15, the TMD forms a trimeric helical bundle with inter-helical distances of 8.2-8.4Å for L493F and L504F and 10.5Å for L500F. These data provide high-resolution evidence of trimer formation of a viral fusion protein TMD in phospholipid bilayers, and indicate that the parainfluenza virus 5 fusion protein TMD harbors two functions: the central α-helical core is the trimerization unit of the protein, while the two termini are responsible for inducing membrane curvature by transitioning to a β-sheet conformation.
Keywords: conformational plasticity; magic-angle-spinning NMR; spin diffusion; trimer formation.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

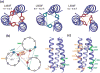
References
-
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

