Targeted Synthesis and Characterization of a Gene Cluster Encoding NAD(P)H-Dependent 3α-, 3β-, and 12α-Hydroxysteroid Dehydrogenases from Eggerthella CAG:298, a Gut Metagenomic Sequence
- PMID: 29330189
- PMCID: PMC5861830
- DOI: 10.1128/AEM.02475-17
Targeted Synthesis and Characterization of a Gene Cluster Encoding NAD(P)H-Dependent 3α-, 3β-, and 12α-Hydroxysteroid Dehydrogenases from Eggerthella CAG:298, a Gut Metagenomic Sequence
Abstract
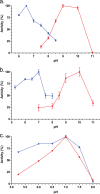
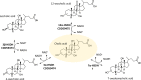
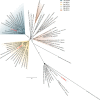
Gut metagenomic sequences provide a rich source of microbial genes, the majority of which are annotated by homology or unknown. Genes and gene pathways that encode enzymes catalyzing biotransformation of host bile acids are important to identify in gut metagenomic sequences due to the importance of bile acids in gut microbiome structure and host physiology. Hydroxysteroid dehydrogenases (HSDHs) are pyridine nucleotide-dependent enzymes with stereospecificity and regiospecificity for bile acid and steroid hydroxyl groups. HSDHs have been identified in several protein families, including medium-chain and short-chain dehydrogenase/reductase families as well as the aldo-keto reductase family. These protein families are large and contain diverse functionalities, making prediction of HSDH-encoding genes difficult and necessitating biochemical characterization. We located a gene cluster in Eggerthella sp. CAG:298 predicted to encode three HSDHs (CDD59473, CDD59474, and CDD59475) and synthesized the genes for heterologous expression in Escherichia coli We then screened bile acid substrates against the purified recombinant enzymes. CDD59475 is a novel 12α-HSDH, and we determined that CDD59474 (3α-HSDH) and CDD59473 (3β-HSDH) constitute novel enzymes in an iso-bile acid pathway. Phylogenetic analysis of these HSDHs with other gut bacterial HSDHs and closest homologues in the database revealed predictable clustering of HSDHs by function and identified several likely HSDH sequences from bacteria isolated or sequenced from diverse mammalian and avian gut samples.IMPORTANCE Bacterial HSDHs have the potential to significantly alter the physicochemical properties of bile acids, with implications for increased/decreased toxicity for gut bacteria and the host. The generation of oxo-bile acids is known to inhibit host enzymes involved in glucocorticoid metabolism and may alter signaling through nuclear receptors such as farnesoid X receptor and G-protein-coupled receptor TGR5. Biochemical or similar approaches are required to fill in many gaps in our ability to link a particular enzymatic function with a nucleic acid or amino acid sequence. In this regard, we have identified a novel 12α-HSDH and a novel set of genes encoding an iso-bile acid pathway (3α-HSDH and 3β-HSDH) involved in epimerization and detoxification of harmful secondary bile acids.
Keywords: Eggerthella; bile acid; hydroxysteroid dehydrogenase; metagenome; targeted gene synthesis.
Copyright © 2018 American Society for Microbiology.
Figures


References
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. - DOI - PMC - PubMed
-
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. doi: 10.1016/j.cmet.2013.01.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

