Effects of allelic variations in the human myxovirus resistance protein A on its antiviral activity
- PMID: 29330299
- PMCID: PMC5836113
- DOI: 10.1074/jbc.M117.812784
Effects of allelic variations in the human myxovirus resistance protein A on its antiviral activity
Abstract
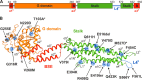
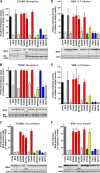
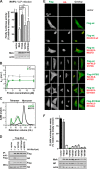
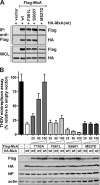
Only a minority of patients infected with seasonal influenza A viruses exhibit a severe or fatal outcome of infection, but the reasons for this inter-individual variability in influenza susceptibility are unclear. To gain further insights into the molecular mechanisms underlying this variability, we investigated naturally occurring allelic variations of the myxovirus resistance 1 (MX1) gene coding for the influenza restriction factor MxA. The interferon-induced dynamin-like GTPase consists of an N-terminal GTPase domain, a bundle signaling element, and a C-terminal stalk responsible for oligomerization and viral target recognition. We used online databases to search for variations in the MX1 gene. Deploying in vitro approaches, we found that non-synonymous variations in the GTPase domain cause the loss of antiviral and enzymatic activities. Furthermore, we showed that these amino acid substitutions disrupt the interface for GTPase domain dimerization required for the stimulation of GTP hydrolysis. Variations in the stalk were neutral or slightly enhanced or abolished MxA antiviral function. Remarkably, two other stalk variants altered MxA's antiviral specificity. Variations causing the loss of antiviral activity were found only in heterozygous carriers. Interestingly, the inactive stalk variants blocked the antiviral activity of WT MxA in a dominant-negative way, suggesting that heterozygotes are phenotypically MxA-negative. In contrast, the GTPase-deficient variants showed no dominant-negative effect, indicating that heterozygous carriers should remain unaffected. Our results demonstrate that naturally occurring mutations in the human MX1 gene can influence MxA function, which may explain individual variations in influenza virus susceptibility in the human population.
Keywords: Mx proteins; allelic variations; antiviral response; dynamin; genetic polymorphism; influenza virus; innate immunity; interferon.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Gao S., von der Malsburg A., Dick A., Faelber K., Schröder G. F., Haller O., Kochs G., and Daumke O. (2011) Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35, 514–525 10.1016/j.immuni.2011.07.012 - DOI - PubMed
-
- Mitchell P. S., Patzina C., Emerman M., Haller O., Malik H. S., and Kochs G. (2012) Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe 12, 598–604 10.1016/j.chom.2012.09.005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

