The RES complex is required for efficient transformation of the precatalytic B spliceosome into an activated Bact complex
- PMID: 29330354
- PMCID: PMC5795787
- DOI: 10.1101/gad.308163.117
The RES complex is required for efficient transformation of the precatalytic B spliceosome into an activated Bact complex
Abstract
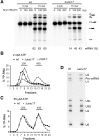
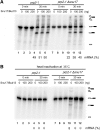
The precise function of the trimeric retention and splicing (RES) complex in pre-mRNA splicing remains unclear. Here we dissected the role of RES during the assembly and activation of yeast spliceosomes. The efficiency of pre-mRNA splicing was significantly lower in the absence of the RES protein Snu17, and the recruitment of its binding partners, Pml1 (pre-mRNA leakage protein 1) and Bud13 (bud site selection protein 13), to the spliceosome was either abolished or substantially reduced. RES was not required for the assembly of spliceosomal B complexes, but its absence hindered efficient Bact complex formation. ΔRES spliceosomes were no longer strictly dependent on Prp2 activity for their catalytic activation, suggesting that they are structurally compromised. Addition of Prp2, Spp2, and UTP to affinity-purified ΔRES B or a mixture of B/Bact complexes formed on wild-type pre-mRNA led to their disassembly. However, no substantial disassembly was observed with ΔRES spliceosomes formed on a truncated pre-mRNA that allows Prp2 binding but blocks its activity. Thus, in the absence of RES, Prp2 appears to bind prematurely, leading to the disassembly of the ΔRES B complexes to which it binds. Our data suggest that Prp2 can dismantle B complexes with an aberrant protein composition, suggesting that it may proofread the spliceosome's RNP structure prior to activation.
Keywords: Prp2 ATPase/RNA helicase; RES complex; pre-mRNA splicing; spliceosome activation; spliceosome disassembly.
© 2018 Bao et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, Stark H. 2017. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
