Tracheophyte genomes keep track of the deep evolution of the Caulimoviridae
- PMID: 29330451
- PMCID: PMC5766536
- DOI: 10.1038/s41598-017-16399-x
Tracheophyte genomes keep track of the deep evolution of the Caulimoviridae
Abstract
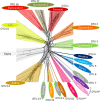
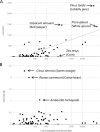
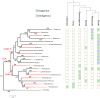
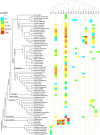
Endogenous viral elements (EVEs) are viral sequences that are integrated in the nuclear genomes of their hosts and are signatures of viral infections that may have occurred millions of years ago. The study of EVEs, coined paleovirology, provides important insights into virus evolution. The Caulimoviridae is the most common group of EVEs in plants, although their presence has often been overlooked in plant genome studies. We have refined methods for the identification of caulimovirid EVEs and interrogated the genomes of a broad diversity of plant taxa, from algae to advanced flowering plants. Evidence is provided that almost every vascular plant (tracheophyte), including the most primitive taxa (clubmosses, ferns and gymnosperms) contains caulimovirid EVEs, many of which represent previously unrecognized evolutionary branches. In angiosperms, EVEs from at least one and as many as five different caulimovirid genera were frequently detected, and florendoviruses were the most widely distributed, followed by petuviruses. From the analysis of the distribution of different caulimovirid genera within different plant species, we propose a working evolutionary scenario in which this family of viruses emerged at latest during Devonian era (approx. 320 million years ago) followed by vertical transmission and by several cross-division host swaps.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

