Supervised Machine Learning for Population Genetics: A New Paradigm
- PMID: 29331490
- PMCID: PMC5905713
- DOI: 10.1016/j.tig.2017.12.005
Supervised Machine Learning for Population Genetics: A New Paradigm
Abstract
As population genomic datasets grow in size, researchers are faced with the daunting task of making sense of a flood of information. To keep pace with this explosion of data, computational methodologies for population genetic inference are rapidly being developed to best utilize genomic sequence data. In this review we discuss a new paradigm that has emerged in computational population genomics: that of supervised machine learning (ML). We review the fundamentals of ML, discuss recent applications of supervised ML to population genetics that outperform competing methods, and describe promising future directions in this area. Ultimately, we argue that supervised ML is an important and underutilized tool that has considerable potential for the world of evolutionary genomics.
Copyright © 2017 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures
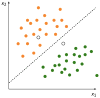

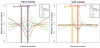
References
-
- Breiman L. Statistical modeling: the two cultures (with comments and a rejoinder by the author) Stat Sci. 2001;16:199–231.
-
- Hinton G, et al. Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Process Mag. 2012;29:82–97.
-
- Sebastiani F. Machine learning in automated text categorization. ACM Comput Surv. 2002;34:1–47.
-
- Krizhevsky A, et al. Imagenet classification with deep convolutional neural networks. In: Fereira F, editor. Advances in Neural Information Processing Systems 25. Neural Information Processing Systems Foundation; 2012. pp. 1097–1105.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

