Transposable elements: genome innovation, chromosome diversity, and centromere conflict
- PMID: 29332159
- PMCID: PMC5857280
- DOI: 10.1007/s10577-017-9569-5
Transposable elements: genome innovation, chromosome diversity, and centromere conflict
Abstract
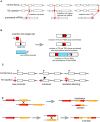
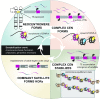
Although it was nearly 70 years ago when transposable elements (TEs) were first discovered "jumping" from one genomic location to another, TEs are now recognized as contributors to genomic innovations as well as genome instability across a wide variety of species. In this review, we illustrate the ways in which active TEs, specifically retroelements, can create novel chromosome rearrangements and impact gene expression, leading to disease in some cases and species-specific diversity in others. We explore the ways in which eukaryotic genomes have evolved defense mechanisms to temper TE activity and the ways in which TEs continue to influence genome structure despite being rendered transpositionally inactive. Finally, we focus on the role of TEs in the establishment, maintenance, and stabilization of critical, yet rapidly evolving, chromosome features: eukaryotic centromeres. Across centromeres, specific types of TEs participate in genomic conflict, a balancing act wherein they are actively inserting into centromeric domains yet are harnessed for the recruitment of centromeric histones and potentially new centromere formation.
Keywords: Centromeric retroelement; Chromosome evolution; Conflict; Genome defense; Satellite; TE; Transposable element.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

