Next-Generation Rapid Autopsies Enable Tumor Evolution Tracking and Generation of Preclinical Models
- PMID: 29333526
- PMCID: PMC5761727
- DOI: 10.1200/PO.16.00038
Next-Generation Rapid Autopsies Enable Tumor Evolution Tracking and Generation of Preclinical Models
Abstract
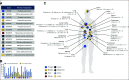
Purpose: Patients with cancer who graciously consent for autopsy represent an invaluable resource for the study of cancer biology. To advance the study of tumor evolution, metastases, and resistance to treatment, we developed a next-generation rapid autopsy program integrated within a broader precision medicine clinical trial that interrogates pre- and postmortem tissue samples for patients of all ages and cancer types.
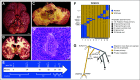
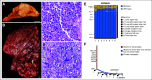
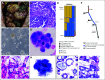
Materials and methods: One hundred twenty-three (22%) of 554 patients who consented to the clinical trial also consented for rapid autopsy. This report comprises the first 15 autopsies, including patients with metastatic carcinoma (n = 10), melanoma (n = 1), and glioma (n = 4). Whole-exome sequencing (WES) was performed on frozen autopsy tumor samples from multiple anatomic sites and on non-neoplastic tissue. RNA sequencing (RNA-Seq) was performed on a subset of frozen samples. Tissue was also used for the development of preclinical models, including tumor organoids and patient-derived xenografts.
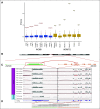
Results: Three hundred forty-six frozen samples were procured in total. WES was performed on 113 samples and RNA-Seq on 72 samples. Successful cell strain, tumor organoid, and/or patient-derived xenograft development was achieved in four samples, including an inoperable pediatric glioma. WES data were used to assess clonal evolution and molecular heterogeneity of tumors in individual patients. Mutational profiles of primary tumors and metastases yielded candidate mediators of metastatic spread and organotropism including CUL9 and PIGM in metastatic ependymoma and ANKRD52 in metastatic melanoma to the lung. RNA-Seq data identified novel gene fusion candidates.
Conclusion: A next-generation sequencing-based autopsy program in conjunction with a pre-mortem precision medicine pipeline for diverse tumors affords a valuable window into clonal evolution, metastasis, and alterations underlying treatment. Moreover, such an autopsy program yields robust preclinical models of disease.
Conflict of interest statement
Next-Generation Rapid Autopsies Enable Tumor Evolution Tracking and Generation of Preclinical Models
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
David J. Pisapia
No relationship to disclose
Steven Salvatore
No relationship to disclose
Chantal Pauli
No relationship to disclose
Erika Hissong
No relationship to disclose
Ken Eng
No relationship to disclose
Davide Prandi
No relationship to disclose
Verena-Wilbeth Sailer
No relationship to disclose
Brian D. Robinson
Kyung Park
No relationship to disclose
Joanna Cyrta
No relationship to disclose
Scott T. Tagawa
Myriam Kossai
No relationship to disclose
Jacqueline Fontugne
No relationship to disclose
Robert Kim
No relationship to disclose
Alexandros Sigaras
Rema Rao
No relationship to disclose
Danielle Pancirer
No relationship to disclose
Bishoy Faltas
No relationship to disclose
Rohan Bareja
No relationship to disclose
Ana M. Molina
David M. Nanus
Prajwal Rajappa
No relationship to disclose
Mark M. Souweidane
Jeffrey Greenfield
No relationship to disclose
Anne-Katrin Emde
No relationship to disclose
Nicolas Robine
No relationship to disclose
Olivier Elemento
No relationship to disclose
Andrea Sboner
No relationship to disclose
Francesca Demichelis
Himisha Beltran
Mark A. Rubin
Juan Miguel Mosquera
No relationship to disclose
Figures

References
-
- Pauli C, Puca L, Mosquera JM, et al. : An emerging role for cytopathology in precision oncology. Cancer Cytopathol 124:167-173, 2016 - PubMed
-
- Rubin MA, Putzi M, Mucci N, et al: Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res 6:1038-1045, 2000. - PubMed
-
- Shah RB, Mehra R, Chinnaiyan AM, et al. : Androgen-independent prostate cancer is a heterogeneous group of diseases: Lessons from a rapid autopsy program. Cancer Res 64:9209-9216, 2004 - PubMed
-
- Zarghooni M, Bartels U, Lee E, et al. : Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol 28:1337-1344, 2010 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

