Dual gene activation and knockout screen reveals directional dependencies in genetic networks
- PMID: 29334369
- PMCID: PMC6072461
- DOI: 10.1038/nbt.4062
Dual gene activation and knockout screen reveals directional dependencies in genetic networks
Abstract
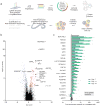
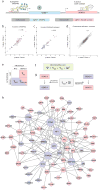
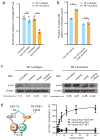
Understanding the direction of information flow is essential for characterizing how genetic networks affect phenotypes. However, methods to find genetic interactions largely fail to reveal directional dependencies. We combine two orthogonal Cas9 proteins from Streptococcus pyogenes and Staphylococcus aureus to carry out a dual screen in which one gene is activated while a second gene is deleted in the same cell. We analyze the quantitative effects of activation and knockout to calculate genetic interaction and directionality scores for each gene pair. Based on the results from over 100,000 perturbed gene pairs, we reconstruct a directional dependency network for human K562 leukemia cells and demonstrate how our approach allows the determination of directionality in activating genetic interactions. Our interaction network connects previously uncharacterized genes to well-studied pathways and identifies targets relevant for therapeutic intervention.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Comment in
-
Pulling the genome in opposite directions to dissect gene networks.Genome Biol. 2018 Mar 26;19(1):42. doi: 10.1186/s13059-018-1425-1. Genome Biol. 2018. PMID: 29580291 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

