Plasma virome of cattle from forest region revealed diverse small circular ssDNA viral genomes
- PMID: 29334978
- PMCID: PMC5769433
- DOI: 10.1186/s12985-018-0923-9
Plasma virome of cattle from forest region revealed diverse small circular ssDNA viral genomes
Abstract
Background: Free-range cattle are common in the Northeast China area, which have close contact with farmers and may carry virus threatening to cattle and farmers.
Methods: Using viral metagenomics we analyzed the virome in plasma samples collected from 80 cattle from the forested region of Northeast China.
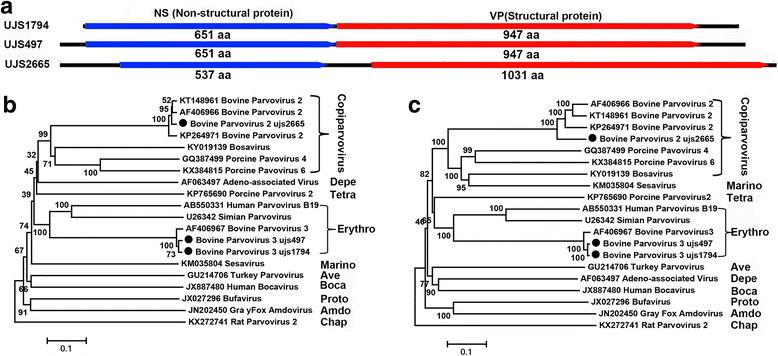
Results: The virome of cattle plasma is composed of the viruses belonging to the families including Parvoviridae, Papillomaviridae, Picobirnaviridae, and divergent viral genomes showing sequence similarity to circular Rep-encoding single stranded (CRESS) DNA viruses. Five such CRESS-DNA genomes were full characterized, with Rep sequences related to circovirus and gemycircularvirus. Three bovine parvoviruses belonging to two different genera were also characterized.
Conclusion: The virome in plasma samples of cattle from the forested region of Northeast China was revealed, which further characterized the diversity of viruses in cattle plasma.
Keywords: CRESS-DNA virus; Cattle blood; Parvovirus; Virome.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

