Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn's disease
- PMID: 29335008
- PMCID: PMC5769311
- DOI: 10.1186/s40168-018-0398-3
Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn's disease
Abstract
Background: Crohn's disease (CD) has an unclear etiology, but there is growing evidence of a direct link with a dysbiotic microbiome. Many gut microbes have previously been associated with CD, but these have mainly been confounded with patients' ongoing treatments. Additionally, most analyses of CD patients' microbiomes have focused on microbes in stool samples, which yield different insights than profiling biopsy samples.
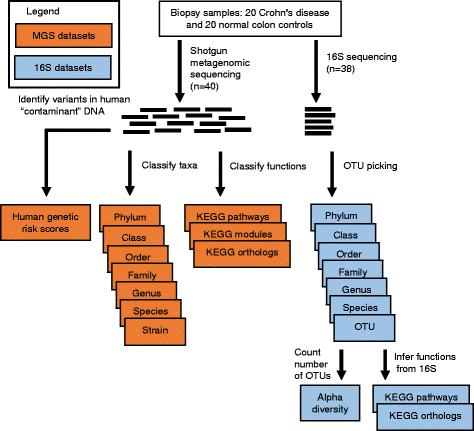
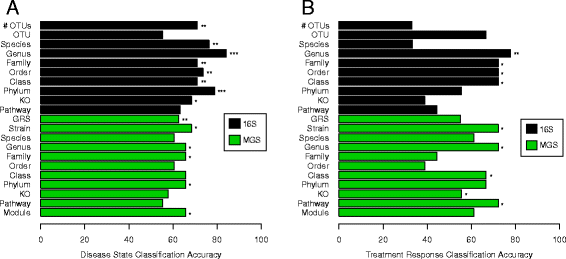
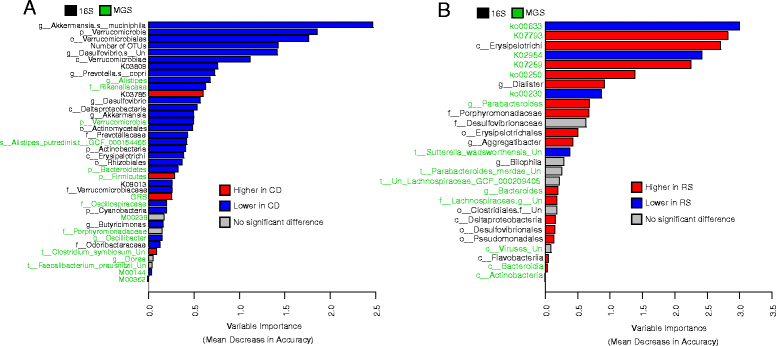
Results: We sequenced the 16S rRNA gene (16S) and carried out shotgun metagenomics (MGS) from the intestinal biopsies of 20 treatment-naïve CD and 20 control pediatric patients. We identified the abundances of microbial taxa and inferred functional categories within each dataset. We also identified known human genetic variants from the MGS data. We then used a machine learning approach to determine the classification accuracy when these datasets, collapsed to different hierarchical groupings, were used independently to classify patients by disease state and by CD patients' response to treatment. We found that 16S-identified microbes could classify patients with higher accuracy in both cases. Based on follow-ups with these patients, we identified which microbes and functions were best for predicting disease state and response to treatment, including several previously identified markers. By combining the top features from all significant models into a single model, we could compare the relative importance of these predictive features. We found that 16S-identified microbes are the best predictors of CD state whereas MGS-identified markers perform best for classifying treatment response.
Conclusions: We demonstrate for the first time that useful predictors of CD treatment response can be produced from shotgun MGS sequencing of biopsy samples despite the complications related to large proportions of host DNA. The top predictive features that we identified in this study could be useful for building an improved classifier for CD and treatment response based on sufferers' microbiome in the future. The BISCUIT project is funded by a Clinical Academic Fellowship from the Chief Scientist Office (Scotland)-CAF/08/01.
Keywords: Crohn’s disease; Machine learning; Microbiome; Pediatric; Treatment response; Treatment-naïve.
Conflict of interest statement
Ethics approval and consent to participate
Ethical approval was granted by North of Scotland Research Ethics Service (09/S0802/24), and written informed consent was obtained from the parents of all subjects. Informed assent was also obtained from older children who were deemed capable of understanding the nature of the study. This study is publicly registered on the United Kingdom Clinical Research Network Portfolio (9633). An ethics amendment allowed further review of the cohort participants’ therapies and clinical response for the first year following diagnosis.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

