Ensemble genomic analysis in human lung tissue identifies novel genes for chronic obstructive pulmonary disease
- PMID: 29335020
- PMCID: PMC5769240
- DOI: 10.1186/s40246-018-0132-z
Ensemble genomic analysis in human lung tissue identifies novel genes for chronic obstructive pulmonary disease
Abstract
Background: Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) significantly associated with chronic obstructive pulmonary disease (COPD). However, many genetic variants show suggestive evidence for association but do not meet the strict threshold for genome-wide significance. Integrative analysis of multiple omics datasets has the potential to identify novel genes involved in disease pathogenesis by leveraging these variants in a functional, regulatory context.
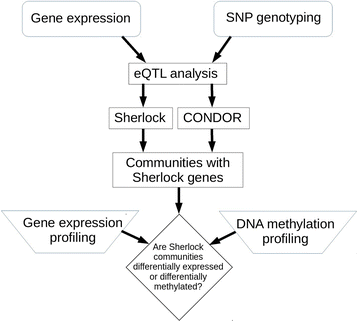
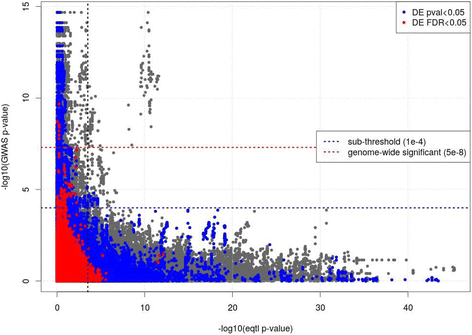
Results: We performed expression quantitative trait locus (eQTL) analysis using genome-wide SNP genotyping and gene expression profiling of lung tissue samples from 86 COPD cases and 31 controls, testing for SNPs associated with gene expression levels. These results were integrated with a prior COPD GWAS using an ensemble statistical and network methods approach to identify relevant genes and observe them in the context of overall genetic control of gene expression to highlight co-regulated genes and disease pathways. We identified 250,312 unique SNPs and 4997 genes in the cis(local)-eQTL analysis (5% false discovery rate). The top gene from the integrative analysis was MAPT, a gene recently identified in an independent GWAS of lung function. The genes HNRNPAB and PCBP2 with RNA binding activity and the gene ACVR1B were identified in network communities with validated disease relevance.
Conclusions: The integration of lung tissue gene expression with genome-wide SNP genotyping and subsequent intersection with prior GWAS and omics studies highlighted candidate genes within COPD loci and in communities harboring known COPD genes. This integration also identified novel disease genes in sub-threshold regions that would otherwise have been missed through GWAS.
Trial registration: ClinicalTrials.gov NCT00608764 NCT00292552.
Keywords: Bayesian methods; Ensemble methods; Expression QTL; Integrative genomics; Network medicine; eQTL.
Conflict of interest statement
Ethics approval and consent to participate
Subjects provided written informed consent for the use of excess lung tissue for research. IRB approval was obtained at Partners Healthcare (parent company of Brigham and Women’s Hospital), Temple University, and St. Elizabeth’s Hospital. The methods for lung tissue research were carried out in accordance with the relevant guidelines.
Competing interests
Drs. Morrow, Platig, Zhou, Qiu, Marchetti, Criner, Celli, Glass, Quackenbush report no competing interests related to this manuscript.
Dr. Cho has received compensation from GSK.
Dr. DeMeo has received compensation from Novartis.
Dr. Bueno has received compensation from Myriad Genetics, Inc., Siemens, Verastem, Inc., Genentech, Inc., Gritstone Oncology, Inc., HTG Molecular Diagnostics, Inc., Neil Leifer, Esq, Morrison Mahoney, David Weiss, LLC, Balick & Balick, LLC, Hartford Hospital, Cleveland Clinic/Conference, Aspen Lung Conference, Case Western Reserve, North Shore University, Castle Bioscience, Novartis Institutes for Biomedical Research, AstraZebeca, imCORE/Roche, Arthur Tuverson, LLC, Ferraro Law Firm, Rice Dolan & Kershaw, Satterly & Kelly, LLC, Exosome, Inc.
Dr. Washko has received compensation from Boehringer Ingelheim, GlaxoSmithKline, Janssen Pharmaceuticals, BTG Interventional Medicine, Regeneron, Quantitative Imaging Solutions and ModoSpira, and his Spouse works for Biogen.
Dr. Silverman has received compensation from COPD Foundation, GlaxoSmithKline, Merck and Novartis,
Dr. Hersh has received consulting fees from AstraZeneca, Concert Pharmaceuticals, Mylan, and grant support from Boehrinher Ingelheim.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Silverman EK. Genetics of chronic obstructive pulmonary disease. In: Organizer DC, Goode JA, editors. Chronic Obstr. Pulm. Dis. Pathog. Treat. John Wiley & Sons, Ltd; 2000. p. 45–64.
-
- Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. doi: 10.1164/ajrccm.157.6.9706014. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- N01 HR076102/HL/NHLBI NIH HHS/United States
- R25 ES011080/ES/NIEHS NIH HHS/United States
- N01 HR076114/HL/NHLBI NIH HHS/United States
- N01 HR076109/HL/NHLBI NIH HHS/United States
- N01 HR076105/HL/NHLBI NIH HHS/United States
- N01 HR076115/HR/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- R01 HL089897/HL/NHLBI NIH HHS/United States
- N01 HR076103/HL/NHLBI NIH HHS/United States
- R01 HL130512/HL/NHLBI NIH HHS/United States
- N01 HR076113/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- N01 HR076104/HL/NHLBI NIH HHS/United States
- K25 HL133599/HL/NHLBI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- N01 HR076101/HL/NHLBI NIH HHS/United States
- N01 HR076110/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- N01 HR076112/HL/NHLBI NIH HHS/United States
- P01 HL105339/HL/NHLBI NIH HHS/United States
- N01 HR076119/HL/NHLBI NIH HHS/United States
- N01 HR076108/HL/NHLBI NIH HHS/United States
- N01 HR076116/HL/NHLBI NIH HHS/United States
- N01 HR076111/HL/NHLBI NIH HHS/United States
- K25 HL136846/HL/NHLBI NIH HHS/United States
- N01 HR076118/HL/NHLBI NIH HHS/United States
- N01 HR076107/HL/NHLBI NIH HHS/United States
- R01 HL125583/HL/NHLBI NIH HHS/United States
- R01 HL127200/HL/NHLBI NIH HHS/United States
- N01 HR076106/HL/NHLBI NIH HHS/United States
- T32 HL007427/HL/NHLBI NIH HHS/United States
- R01 HL111759/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

