Fungi stabilize connectivity in the lung and skin microbial ecosystems
- PMID: 29335027
- PMCID: PMC5769346
- DOI: 10.1186/s40168-017-0393-0
Fungi stabilize connectivity in the lung and skin microbial ecosystems
Abstract
Background: No microbe exists in isolation, and few live in environments with only members of their own kingdom or domain. As microbiome studies become increasingly more interested in the interactions between microbes than in cataloging which microbes are present, the variety of microbes in the community should be considered. However, the majority of ecological interaction networks for microbiomes built to date have included only bacteria. Joint association inference across multiple domains of life, e.g., fungal communities (the mycobiome) and bacterial communities, has remained largely elusive.
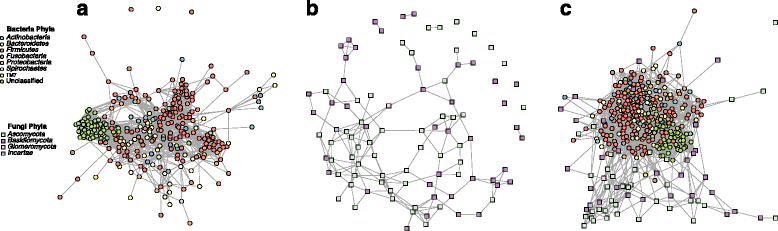
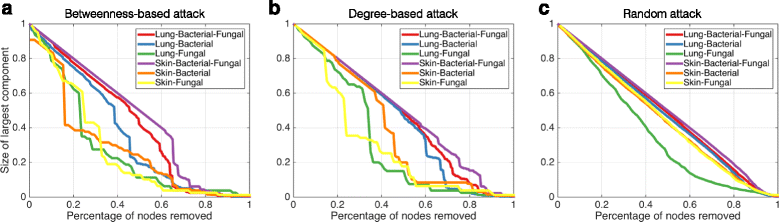
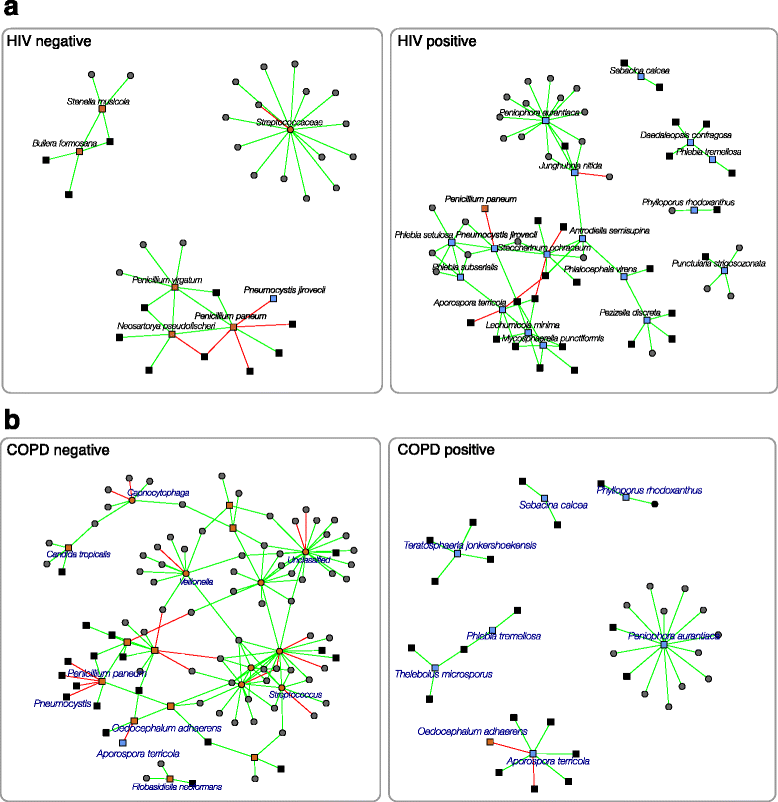
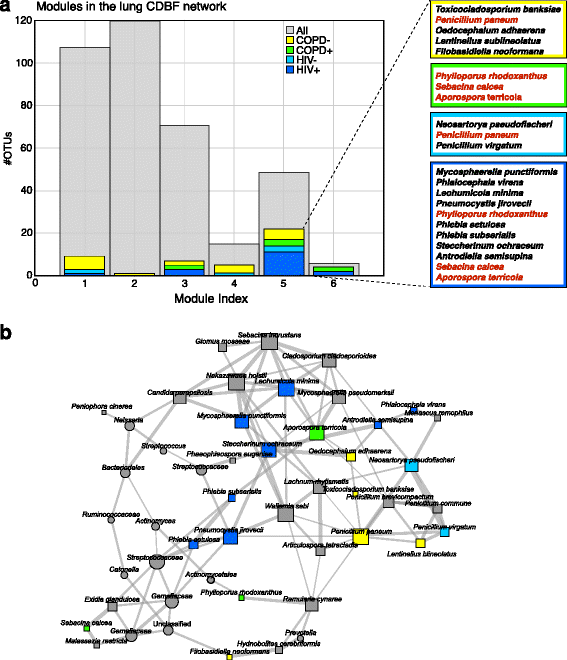
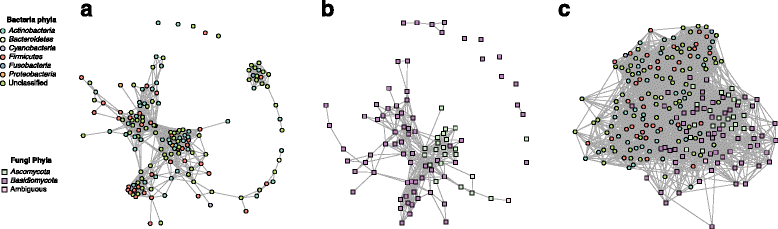
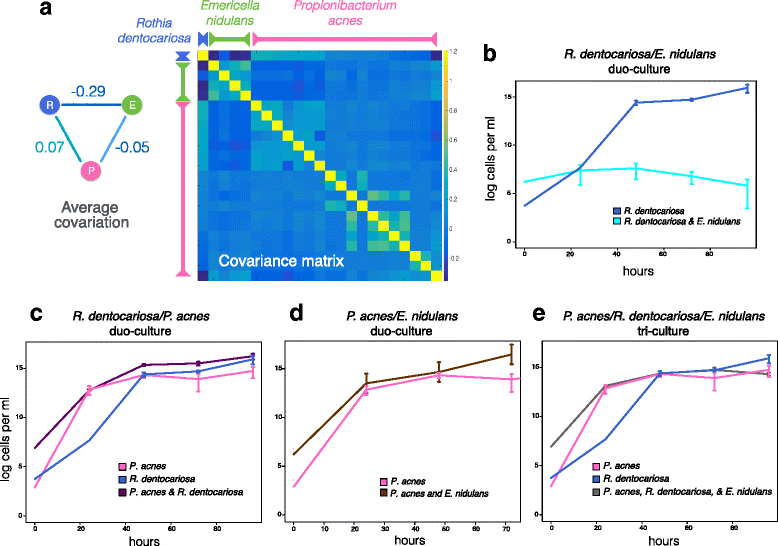
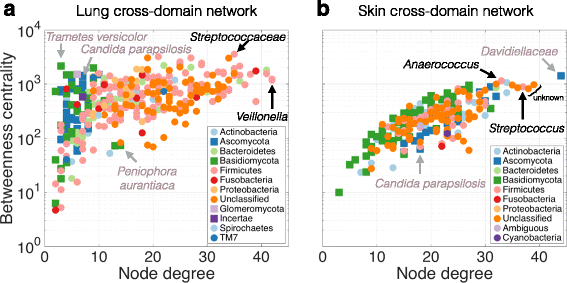
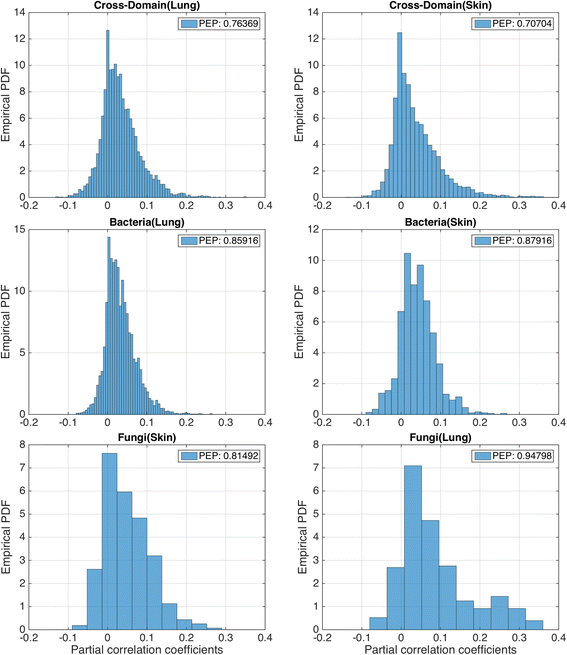
Results: Here, we present a novel extension of the SParse InversE Covariance estimation for Ecological ASsociation Inference (SPIEC-EASI) framework that allows statistical inference of cross-domain associations from targeted amplicon sequencing data. For human lung and skin micro- and mycobiomes, we show that cross-domain networks exhibit higher connectivity, increased network stability, and similar topological re-organization patterns compared to single-domain networks. We also validate in vitro a small number of cross-domain interactions predicted by the skin association network.
Conclusions: For the human lung and skin micro- and mycobiomes, our findings suggest that fungi play a stabilizing role in ecological network organization. Our study suggests that computational efforts to infer association networks that include all forms of microbial life, paired with large-scale culture-based association validation experiments, will help formulate concrete hypotheses about the underlying biological mechanisms of species interactions and, ultimately, help understand microbial communities as a whole.
Conflict of interest statement
Ethics approval and consent to participate
Written informed consent was obtained from all participants in both studies following approval of human subjects’ protection protocols from review boards of the University of Pittsburgh, University of California San Francisco, the University of California Los Angeles, and the National Human Genetics Research Institute.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- K24 HL123342/HL/NHLBI NIH HHS/United States
- PN2 EY016586/EY/NEI NIH HHS/United States
- U01 HL098962/HL/NHLBI NIH HHS/United States
- GM32877-21/22/National Institutes of Health (US)/International
- EY016586-06/NH/NIH HHS/United States
- U54 CA143907/CA/NCI NIH HHS/United States
- K24 HL123342/NH/NIH HHS/United States
- PN2-EY016586/NH/NIH HHS/United States
- U01HL098962/National Institutes of Health (US)/International
- IU54CA143907-01/NH/NIH HHS/United States
- K24 HL087713/HL/NHLBI NIH HHS/United States
- IOS-1126971/National Science Foundation/International
- R01 HL090339/HL/NHLBI NIH HHS/United States
- R01 GM032877/GM/NIGMS NIH HHS/United States
- PN1 EY016586/EY/NEI NIH HHS/United States
- R01HL090339/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

