Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing
- PMID: 29335443
- PMCID: PMC5768770
- DOI: 10.1038/s41467-017-02584-z
Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing
Abstract
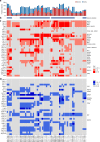
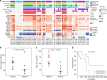
EGFR-mutant lung adenocarcinomas (LUAD) display diverse clinical trajectories and are characterized by rapid but short-lived responses to EGFR tyrosine kinase inhibitors (TKIs). Through sequencing of 79 spatially distinct regions from 16 early stage tumors, we show that despite low mutation burdens, EGFR-mutant Asian LUADs unexpectedly exhibit a complex genomic landscape with frequent and early whole-genome doubling, aneuploidy, and high clonal diversity. Multiple truncal alterations, including TP53 mutations and loss of CDKN2A and RB1, converge on cell cycle dysregulation, with late sector-specific high-amplitude amplifications and deletions that potentially beget drug resistant clones. We highlight the association between genomic architecture and clinical phenotypes, such as co-occurring truncal drivers and primary TKI resistance. Through comparative analysis with published smoking-related LUAD, we postulate that the high intra-tumor heterogeneity observed in Asian EGFR-mutant LUAD may be contributed by an early dominant driver, genomic instability, and low background mutation rates.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

