Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis
- PMID: 29335544
- PMCID: PMC5805582
- DOI: 10.1038/s41588-017-0032-5
Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis
Abstract
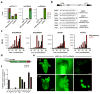
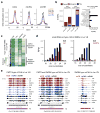
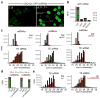
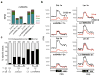
Chromosome dosage has substantial effects on reproductive isolation and speciation in both plants and animals, but the underlying mechanisms are largely obscure 1 . Transposable elements in animals can regulate hybridity through maternal small RNA 2 , whereas small RNAs in plants have been postulated to regulate dosage response via neighboring imprinted genes3,4. Here we show that a highly conserved microRNA in plants, miR845, targets the tRNAMet primer-binding site (PBS) of long terminal repeat (LTR) retrotransposons in Arabidopsis pollen, and triggers the accumulation of 21-22-nucleotide (nt) small RNAs in a dose-dependent fashion via RNA polymerase IV. We show that these epigenetically activated small interfering RNAs (easiRNAs) mediate hybridization barriers between diploid seed parents and tetraploid pollen parents (the 'triploid block'), and that natural variation for miR845 may account for 'endosperm balance' allowing the formation of triploid seeds. Targeting of the PBS with small RNA is a common mechanism for transposon control in mammals and plants, and provides a uniquely sensitive means to monitor chromosome dosage and imprinting in the developing seed.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Köhler C, Mittelsten Scheid O, Erilova A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010;26:142–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

