Metatranscriptome of human faecal microbial communities in a cohort of adult men
- PMID: 29335555
- PMCID: PMC6557121
- DOI: 10.1038/s41564-017-0084-4
Metatranscriptome of human faecal microbial communities in a cohort of adult men
Abstract
The gut microbiome is intimately related to human health, but it is not yet known which functional activities are driven by specific microorganisms' ecological configurations or transcription. We report a large-scale investigation of 372 human faecal metatranscriptomes and 929 metagenomes from a subset of 308 men in the Health Professionals Follow-Up Study. We identified a metatranscriptomic 'core' universally transcribed over time and across participants, often by different microorganisms. In contrast to the housekeeping functions enriched in this core, a 'variable' metatranscriptome included specialized pathways that were differentially expressed both across participants and among microorganisms. Finally, longitudinal metagenomic profiles allowed ecological interaction network reconstruction, which remained stable over the six-month timespan, as did strain tracking within and between participants. These results provide an initial characterization of human faecal microbial ecology into core, subject-specific, microorganism-specific and temporally variable transcription, and they differentiate metagenomically versus metatranscriptomically informative aspects of the human faecal microbiome.
Conflict of interest statement
Competing interests
The authors declare no competing financial interests.
Figures


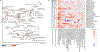


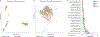
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

