N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications
- PMID: 29335608
- PMCID: PMC6317335
- DOI: 10.1038/s41593-017-0057-1
N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications
Erratum in
-
Publisher Correction: N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications.Nat Neurosci. 2018 Aug;21(8):1139. doi: 10.1038/s41593-018-0169-2. Nat Neurosci. 2018. PMID: 29880878
Abstract
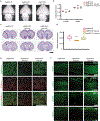
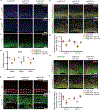
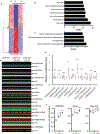
Internal N6-methyladenosine (m6A) modification is widespread in messenger RNAs (mRNAs) and is catalyzed by heterodimers of methyltransferase-like protein 3 (Mettl3) and Mettl14. To understand the role of m6A in development, we deleted Mettl14 in embryonic neural stem cells (NSCs) in a mouse model. Phenotypically, NSCs lacking Mettl14 displayed markedly decreased proliferation and premature differentiation, suggesting that m6A modification enhances NSC self-renewal. Decreases in the NSC pool led to a decreased number of late-born neurons during cortical neurogenesis. Mechanistically, we discovered a genome-wide increase in specific histone modifications in Mettl14 knockout versus control NSCs. These changes correlated with altered gene expression and observed cellular phenotypes, suggesting functional significance of altered histone modifications in knockout cells. Finally, we found that m6A regulates histone modification in part by destabilizing transcripts that encode histone-modifying enzymes. Our results suggest an essential role for m6A in development and reveal m6A-regulated histone modifications as a previously unknown mechanism of gene regulation in mammalian cells.
Figures




References
-
- Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–6 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

