Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression
- PMID: 29337183
- PMCID: PMC6425486
- DOI: 10.1016/j.stem.2017.12.004
Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression
Abstract
Tissue regeneration relies on resident stem cells (SCs), whose activity and lineage choices are influenced by the microenvironment. Exploiting the synchronized, cyclical bouts of tissue regeneration in hair follicles (HFs), we investigate how microenvironment dynamics shape the emergence of stem cell lineages. Employing epigenetic and ChIP-seq profiling, we uncover how signal-dependent transcription factors couple spatiotemporal cues to chromatin dynamics, thereby choreographing stem cell lineages. Using enhancer-driven reporters, mutagenesis, and genetics, we show that simultaneous BMP-inhibitory and WNT signals set the stage for lineage choices by establishing chromatin platforms permissive for diversification. Mechanistically, when binding of BMP effector pSMAD1 is relieved, enhancers driving HF-stem cell master regulators are silenced. Concomitantly, multipotent, lineage-fated enhancers silent in HF-stem cells become activated by exchanging WNT effectors TCF3/4 for LEF1. Throughout regeneration, lineage enhancers continue reliance upon LEF1 but then achieve specificity by accommodating additional incoming signaling effectors. Barriers to progenitor plasticity increase when diverse, signal-sensitive transcription factors shape LEF1-regulated enhancer dynamics.
Keywords: WNT signaling; adult tissue regeneration; chromatin remodeling; epicenter; hair follicle; multipotent progenitors; signaling effectors; stem cell lineage choices; super-enhancer; transient amplifying cells.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures
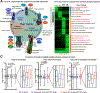
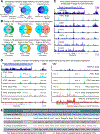


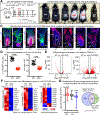
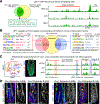
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

