Zinc transporter Slc39a8 is essential for cardiac ventricular compaction
- PMID: 29337306
- PMCID: PMC5785267
- DOI: 10.1172/JCI96993
Zinc transporter Slc39a8 is essential for cardiac ventricular compaction
Abstract
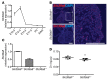
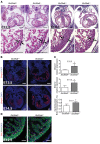
Isolated left ventricular noncompaction (LVNC) results from excessive trabeculation and impaired myocardial compaction during heart development. The extracellular matrix (ECM) that separates endocardium from myocardium plays a critical but poorly understood role in ventricular trabeculation and compaction. In an attempt to characterize solute carrier family 39 member 8-null (Slc39a8-null) mice, we discovered that homozygous null embryos do not survive embryogenesis and exhibit a cardiac phenotype similar to human LVNC. Slc39a8 encodes a divalent metal cation importer that has been implicated in ECM degradation through the zinc/metal regulatory transcription factor 1 (Zn/MTF1) axis, which promotes the expression of ECM-degrading enzymes, including Adamts metalloproteinases. Here, we have shown that Slc39a8 is expressed by endothelial cells in the developing mouse heart, where it serves to maintain cellular Zn levels. Furthermore, Slc39a8-null hearts exhibited marked ECM accumulation and reduction of several Adamts metalloproteinases. Consistent with the in vivo observations, knockdown of SLC39A8 in HUVECs decreased ADAMTS1 transcription by decreasing cellular Zn uptake and, as a result, MTF1 transcriptional activity. Our study thus identifies a gene underlying ventricular trabeculation and compaction development, and a pathway regulating ECM during myocardial morphogenesis.
Keywords: Cardiology; Development; Extracellular matrix; Genetic diseases; Mouse models.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

