Network-Based Methods for Identifying Key Active Proteins in the Extracellular Electron Transfer Process in Shewanella oneidensis MR-1
- PMID: 29337910
- PMCID: PMC5793192
- DOI: 10.3390/genes9010041
Network-Based Methods for Identifying Key Active Proteins in the Extracellular Electron Transfer Process in Shewanella oneidensis MR-1
Abstract
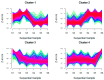
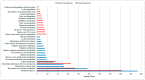
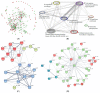
Shewanella oneidensis MR-1 can transfer electrons from the intracellular environment to the extracellular space of the cells to reduce the extracellular insoluble electron acceptors (Extracellular Electron Transfer, EET). Benefiting from this EET capability, Shewanella has been widely used in different areas, such as energy production, wastewater treatment, and bioremediation. Genome-wide proteomics data was used to determine the active proteins involved in activating the EET process. We identified 1012 proteins with decreased expression and 811 proteins with increased expression when the EET process changed from inactivation to activation. We then networked these proteins to construct the active protein networks, and identified the top 20 key active proteins by network centralization analysis, including metabolism- and energy-related proteins, signal and transcriptional regulatory proteins, translation-related proteins, and the EET-related proteins. We also constructed the integrated protein interaction and transcriptional regulatory networks for the active proteins, then found three exclusive active network motifs involved in activating the EET process-Bi-feedforward Loop, Regulatory Cascade with a Feedback, and Feedback with a Protein-Protein Interaction (PPI)-and identified the active proteins involved in these motifs. Both enrichment analysis and comparative analysis to the whole-genome data implicated the multiheme c-type cytochromes and multiple signal processing proteins involved in the process. Furthermore, the interactions of these motif-guided active proteins and the involved functional modules were discussed. Collectively, by using network-based methods, this work reported a proteome-wide search for the key active proteins that potentially activate the EET process.
Keywords: active protein; extracellular electron transfer; network-based methods; protein–protein interaction; transcriptional regulatory interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

