Clustering gene expression time series data using an infinite Gaussian process mixture model
- PMID: 29337990
- PMCID: PMC5786324
- DOI: 10.1371/journal.pcbi.1005896
Clustering gene expression time series data using an infinite Gaussian process mixture model
Abstract
Transcriptome-wide time series expression profiling is used to characterize the cellular response to environmental perturbations. The first step to analyzing transcriptional response data is often to cluster genes with similar responses. Here, we present a nonparametric model-based method, Dirichlet process Gaussian process mixture model (DPGP), which jointly models data clusters with a Dirichlet process and temporal dependencies with Gaussian processes. We demonstrate the accuracy of DPGP in comparison to state-of-the-art approaches using hundreds of simulated data sets. To further test our method, we apply DPGP to published microarray data from a microbial model organism exposed to stress and to novel RNA-seq data from a human cell line exposed to the glucocorticoid dexamethasone. We validate our clusters by examining local transcription factor binding and histone modifications. Our results demonstrate that jointly modeling cluster number and temporal dependencies can reveal shared regulatory mechanisms. DPGP software is freely available online at https://github.com/PrincetonUniversity/DP_GP_cluster.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

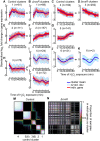


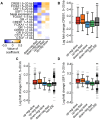
References
-
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293(5537): 2087–2092. doi: 10.1126/science.1061603 - DOI - PubMed
-
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, et al. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297(5590): 2270–2275. doi: 10.1126/science.1072152 - DOI - PubMed
-
- Frank CL, Liu F, Wijayatunge R, Song L, Biegler MT, Yang MG, et al. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015;18(5):647–656. doi: 10.1038/nn.3995 - DOI - PMC - PubMed
-
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12): 4241–4257. doi: 10.1091/mbc.11.12.4241 - DOI - PMC - PubMed
-
- Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2(1): 65–73. doi: 10.1016/S1097-2765(00)80114-8 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

